BackgroundOn March 30, 2013, a novel avian influenza A H7N9 virus that infects human beings was identified. This virus had been detected in six provinces and municipal cities in China as of April 18, 2013. We correlated genomic sequences from avian influenza viruses with ecological information and did phylogenetic and coalescent analyses to extrapolate the potential origins of the virus and possible routes of reassortment events.MethodsWe downloaded H7N9 virus genome sequences from the Global Initiative on Sharing Avian Influenza Data (GISAID) database and public sequences used from the Influenza Virus Resource. We constructed phylogenetic trees and did 1000 bootstrap replicates for each tree. Two rounds of phylogenetic analyses were done. We used at least 100 closely related sequences for each gene to infer the overall topology, removed suspicious sequences from the trees, and focused on the closest clades to the novel H7N9 viruses. We compared our tree topologies with those from a bayesian evolutionary analysis by sampling trees (BEAST) analysis. We used the bayesian Markov chain Monte Carlo method to jointly estimate phylogenies, divergence times, and other evolutionary parameters for all eight gene fragments. We used sequence alignment and homology-modelling methods to study specific mutations regarding phenotypes, specifically addressing the human receptor binding properties.FindingsThe novel avian influenza A H7N9 virus originated from multiple reassortment events. The HA gene might have originated from avian influenza viruses of duck origin, and the NA gene might have transferred from migratory birds infected with avian influenza viruses along the east Asian flyway. The six internal genes of this virus probably originated from two different groups of H9N2 avian influenza viruses, which were isolated from chickens. Detailed analyses also showed that ducks and chickens probably acted as the intermediate hosts leading to the emergence of this virulent H7N9 virus. Genotypic and potential phenotypic differences imply that the isolates causing this outbreak form two separate subclades.InterpretationThe novel avian influenza A H7N9 virus might have evolved from at least four origins. Diversity among isolates implies that the H7N9 virus has evolved into at least two different lineages. Unknown intermediate hosts involved might be implicated, extensive global surveillance is needed, and domestic-poultry-to-person transmission should be closely watched in the future.FundingChina Ministry of Science and Technology Project 973, National Natural Science Foundation of China, China Health and Family Planning Commission, Chinese Academy of Sciences.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
A few things the Symbol Research team are reading. Complex Insight is curated by Phillip Trotter (www.linkedin.com/in/phillip-trotter) from Symbol Research
Curated by
Phillip Trotter
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|




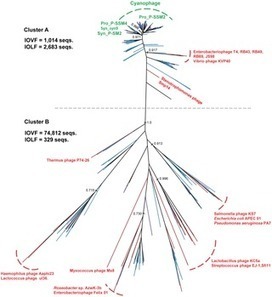





Firstly sidestepping the important findings for H7N9 virus, this paper illustrates the importance of rgorous methodology and key research methods for understanding disease evolution and contagence pathways. the paper details correlated genomic sequences and ecological information using phylogenetic and coalescent analyses to extrapolate the potential originsand possible routes of reassortment events in H7N9 virus. As for the findings - novel avian influenza viruses are a major concern for world wide public health - the research work in this paper raises the need for understanding intermediate hosts, viral evolution pathways and domestic poultry wild animation contact on a global scale for future health policy. Worth reading.