 Your new post is loading...
 Your new post is loading...

|
Scooped by
NatProdChem
|
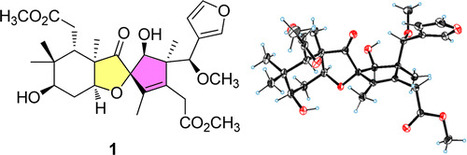
Microorganisms use chemical inactivation strategies to circumvent toxicity caused by many types of antibiotics. Yet in all reported cases, this approach is limited to enzymatically facilitated mechanisms that each target narrow ranges of chemically related scaffolds. The fungus-derived shikimate analogues, pericoxide and pericosine A, were identified as chemoreactive natural products that attenuate the antagonistic effects of several synthetic and naturally derived antifungal agents. Experimental and computational studies suggest that pericoxide and pericosine A readily react via SN2′ mechanisms against a variety of nucleophilic substances under both in vitro aqueous and in situ co-culture conditions. Many of the substitution products from this reaction were highly stable and exhibited diminished toxicities against environmental fungal isolates, including the Tolypocladium sp. strain that produced pericoxide and pericosine A. - Dr. Lin Du†,
- Dr. Jianlan You†,
- Prof. Dr. Kenneth M. Nicholas and
- Prof. Dr. Robert H. Cichewicz*
Article first published online: 1 MAR 2016 DOI: 10.1002/ange.201511348Very interesting

|
Scooped by
NatProdChem
|
Background
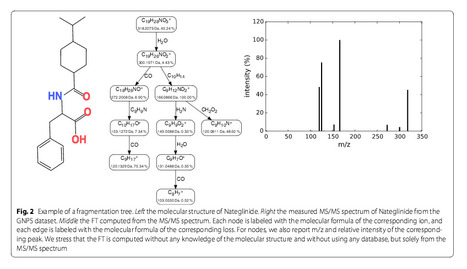
Untargeted metabolomics commonly uses liquid chromatography mass spectrometB¨ry to measure abundances of metabolites; subsequent tandem mass spectrometry is used to derive information about individual compounds. One of the bottlenecks in this experimental setup is the interpretation of fragmentation spectra to accurately and efficiently identify compounds. Fragmentation trees have become a powerful tool for the interpretation of tandem mass spectrometry data of small molecules. These trees are determined from the data using combinatorial optimization, and aim at explaining the experimental data via fragmentation cascades. Fragmentation tree computation does not require spectral or structural databases. To obtain biochemically meaningful trees, one needs an elaborate optimization function (scoring). Results We present a new scoring for computing fragmentation trees, transforming the combinatorial optimization into a Maximum A Posteriori estimator. We demonstrate the superiority of the new scoring for two tasks: both for the de novo identification of molecular formulas of unknown compounds, and for searching a database for structurally similar compounds, our method SIRIUS 3, performs significantly better than the previous version of our method, as well as other methods for this task. Conclusion
SIRIUS 3 can be a part of an untargeted metabolomics workflow, allowing researchers to investigate unknowns using automated computational methods Sebastian BöckerEmail author and Kai Dührkop Journal of Cheminformatics20168:5 DOI: 10.1186/s13321-016-0116-8

|
Scooped by
NatProdChem
|
TAspterpenacids A and B, Two Sesterterpenoids from a Mangrove Endophytic Fungus Aspergillus terreus H010wo new sesterterpenoids, aspterpenacids A (1) and B (2), with an unusual carbon skeleton of a 5/3/7/6/5 ring system were isolated from the mangrove endophytic fungus Aspergillus terreus H010. Their structures were elucidated on the basis of spectroscopic methods, single-crystal X-ray diffraction analysis, and electronic circular dichroism calculations. A biogenetic pathway for 1 and 2 is proposed. Both 1 and 2 showed no significant antibacterial activity or cytotoxicity at 50 μM. Org. Lett., Article ASAP DOI: 10.1021/acs.orglett.6b00336

|
Scooped by
NatProdChem
|
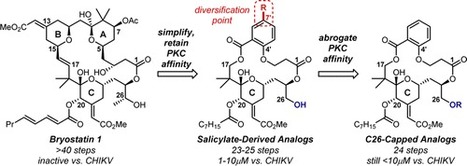
Chikungunya virus (CHIKV) has been spreading rapidly, with over one million confirmed or suspected cases in the Americas since late 2013. Infection with CHIKV causes devastating arthritic and arthralgic symptoms. Currently, there is no therapy to treat this disease, and the only medications focus on relief of symptoms. Recently, protein kinase C (PKC) modulators have been reported to inhibit CHIKV-induced cell death in cell assays. The salicylate-derived bryostatin analogues described here are structurally simplified PKC modulators that are more synthetically accessible than the natural product bryostatin 1, a PKC modulator and clinical lead for the treatment of cancer, Alzheimer’s disease, and HIV eradication. Evaluation of the anti-CHIKV activity of these salicylate-derived bryostatin analogues in cell culture indicates that they are among the most potent cell-protective agents reported to date. Given that they are more accessible and significantly more active than the parent natural product, they represent new therapeutic leads for controlling CHIKV infection. Significantly, these analogues also provide evidence for the involvement of a PKC-independent pathway. This adds a fundamentally distinct aspect to the importance or involvement of PKC modulation in inhibition of chikungunya virus replication, a topic of recent and growing interest.
J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b01017

|
Scooped by
NatProdChem
|
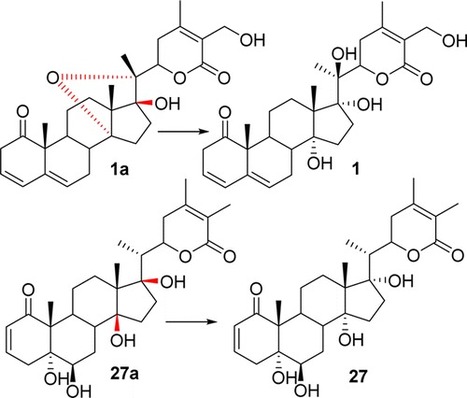
A classic withanolide is defined as a highly oxygenated C28 ergostane-type steroid that is characterized by a C22-hydroxy-C26-oic acid δ-lactone in the nine-carbon side chain. Analysis of the reported 13C NMR data of classic withanolides with hydroxy groups (C-14, C-17, and C-20) revealed that (1) a hydroxy (C-14 or C-17) substituent significantly alters the chemical shifts (C-7, C-9, C-12, and C-21) via the γ-gauche effect; (2) the chemical shift values (C-9, C-12, and C-21) reflect the orientation (α or β) of the hydroxy moiety (C-14 or C-17); (3) a double-bond positional change in ring A (Δ2 to Δ3), or hydroxylation (C-27), results in a minuscule effect on the chemical shifts of carbons in rings C and D (from C-12 to C-18); and (4) the 13C NMR γ-gauche effect method is more convenient and reliable than the traditional approach (1H NMR shift comparisons in C5D5N versus CDCl3) to probe the orientation of the hydroxy substituent (C-14 and C-17). Utilization of these rules demonstrated that the reported 13C NMR data of withanolides 1a–29a were inconsistent with their published structures, which were subsequently revised as 1–16 and 12 and 18–29, respectively. When combined, this strongly supports the application of these methods to determine the relative configuration of steroidal substituents.
Department of Medicinal Chemistry, University of Kansas, Lawrence, Kansas 66045, United States J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b00648 Publication Date (Web): February 19, 2016

|
Scooped by
NatProdChem
|
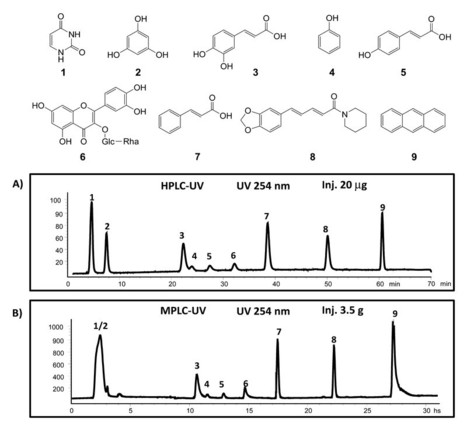
« In natural product research, the isolation of biomarkers or bioactive compounds from complex natural extracts represents an essential step for de novo identification and bioactivity assessment. When pure natural products have to be obtained in milligram quantities, the chromatographic steps are generally labourious and time-consuming. In this respect, an efficient method has been developed for the reversed-phase gradient transfer from high-performance liquid chromatography to medium-performance liquid chromatography for the isolation of pure natural products at the level of tens of milligrams from complex crude natural extracts. The proposed method provides a rational way to predict retention behaviour and resolution at the analytical scale prior to medium-performance liquid chromatography, and guarantees similar performances at both analytical and preparative scales. The optimisation of the high-performance liquid chromatography separation and system characterisation allows for the prediction of the gradient at the medium-performance liquid chromatography scale by using identical stationary phase chemistries. The samples were introduced in medium-performance liquid chromatography using a pressure-resistant aluminium dry load cell especially designed for this study to allow high sample loading while maintaining a maximum achievable flow rate for the separation. The method has been validated with a mixture of eight natural product standards. Ultraviolet and evaporative light scattering detections were used in parallel for a comprehensive monitoring. In addition, post-chromatographic mass spectrometry detection was provided by high-throughput ultrahigh-performance liquid chromatography time-of-flight mass spectrometry analyses of all fractions. The processing of all liquid chromatography-mass spectrometry data in the form of an medium-performance liquid chromatography x ultra high-performance liquid chromatography time-of-flight mass spectrometry matrix enabled an efficient localisation of the compounds of interest in the generated fractions. The methodology was successfully applied for the separation of three different plant extracts that contain many diverse secondary metabolites. The advantages and limitations of this approach and the theoretical chromatographic background that rules such as liquid chromatography gradient transfer are presented from a practical viewpoint. »
Planta Med 2015; 81(17): 1636-1643
DOI: 10.1055/s-0035-1545912 Original Papers Rational and Efficient Preparative Isolation of Natural Products by MPLC-UV-ELSD based on HPLC to MPLC Gradient Transfer Soura Challal1, Emerson Ferreira Queiroz1, Benjamin Debrus1, Werner Kloeti1, Davy Guillarme1, Mahabir Prashad Gupta2, Jean-Luc Wolfender1- 1School of Pharmaceutical Sciences, EPGL, University of Geneva, University of Lausanne, Geneva, Switzerland
- 2Center for Pharmacognostic Research on Panamanian Flora-CIFLORPAN, University of Panama, Panama, Republic of Panama
© 2016 Georg Thieme Verlag KG

|
Scooped by
NatProdChem
|
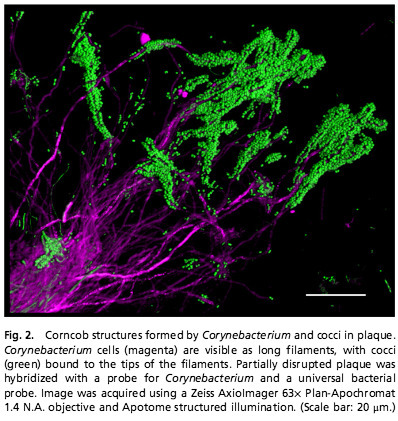
The physiology and ecology of complex microbial communities are strongly dependent on the immediate surroundings of each microbe, including the identity of neighboring microbes; however, information on the micron-scale organization of microbiomes is largely lacking. Using sequencing data combined with spectral fluorescence imaging, we have discovered a multigenus, highly organized microbial consortium in human dental plaque. The spatial structure of the consortium reveals unanticipated interactions and provides a framework for understanding the organization, metabolism, and systems biology of the microbiome and ultimately, its effect on the health of the human host. Our synthesis of high-throughput sequencing data with spatial and structural information shows the informative value of microbial biogeography at the micron scale.
- Jessica L. Mark Welcha,b,1,
- Blair J. Rossettia,b,
- Christopher W. Riekenb,
- Floyd E. Dewhirsta,c, and
- Gary G. Borisya,b,1
PNAS, E791–E800, doi: 10.1073/pnas.1522149113

|
Scooped by
NatProdChem
|
Read the latest article version by Sean Ekins, Daniel Mietchen, Megan Coffee, Thomas P Stratton, Joel S Freundlich, Lucio Freitas-Junior, Eugene Muratov, Jair Siqueira-Neto, Antony J Williams, Carolina Andrade, at F1000Research.

|
Scooped by
NatProdChem
|
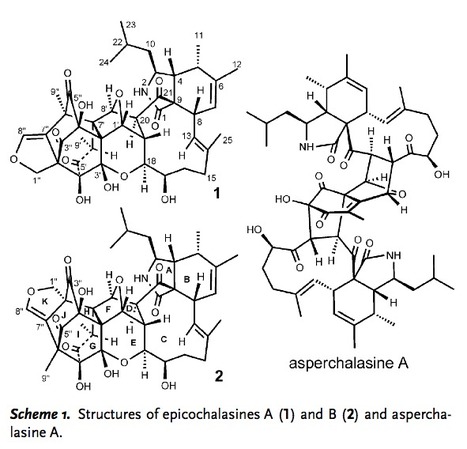
Two bioactive merocytochalasans, epicochalasines A (1) and B (2), a new class of cytochalasans bearing unexpected scaffolds consisting of fused aspochalasin and epicoccine dimer moieties, were isolated from the liquid culture broth of Aspergillus flavipes. Both 1 and 2 possess a hendecacyclic 5/6/11/5/6/5/6/5/6/6/5 ring system containing an adamantyl cage and as many as 19 stereogenic centers; however, the fusion patterns of 1 and 2 differ greatly, thus resulting in different carbon skeletons. The absolute configurations of 1 and 2 were determined by X-ray diffraction and calculated ECD, respectively. The biogenetic pathways of 1 and 2 are proposed to involve Diels–Alder and nucleophilic addition reactions. Both 1 and 2 induced significant G2/M-phase cell-cycle arrest. Furthermore, we found that merocytochalasans induce apoptosis in leukemia cells through the activation of caspase-3 and the degradation of PARP.

|
Scooped by
NatProdChem
|
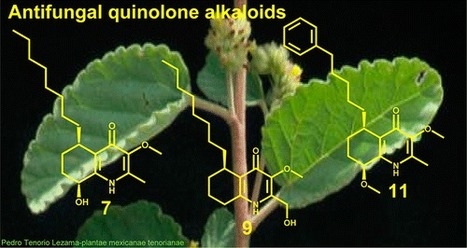
Chemical investigation of a dichloromethane extract of the aerial parts of Waltheria indica led to the isolation and characterization of five polyhydroxymethoxyflavonoids, namely, oxyanin A (1), vitexicarpin (3), chrysosplenol E (4), flindulatin (5), 5-hydroxy-3,7,4′-trimethoxyflavone (6), and six quinolone alkaloids, waltheriones M–Q (2, 7, 8, 10, 11) and 5(R)-vanessine (9). Among these, compounds 2, 7, 8, 10, and 11 have not yet been described in the literature. Their chemical structures were established by means of spectroscopic data interpretation including 1H and 13C, HSQC, HMBC, COSY, and NOESY NMR experiments and UV, IR, and HRESIMS. The absolute configurations of the compounds were established by ECD. The isolated constituents and 10 additional quinoline alkaloids previously isolated from the roots of the plant were evaluated for their in vitro antifungal activity against the human fungal pathogen Candida albicans, and 10 compounds (7, 9, 11–16, 18, 21) showed growth inhibitory activity on both planktonic cells and biofilms (MIC ≤ 32 μg/mL). Their spectrum of activity against other pathogenic Candida species and their cytotoxicity against human HeLa cells were also determined. In addition, the cytological effect of the antifungal isolated compounds on the ultrastructure of C. albicans was evaluated by transmission electron microscopy.
Sylvian Cretton†, Stéphane Dorsaz‡, Antonio Azzollini†, Quentin Favre-Godal†, Laurence Marcourt†, Samad Nejad Ebrahimi§, Francine Voinesco⊥, Emilie Michellod⊥, Dominique Sanglard‡, Katia Gindro⊥, Jean-Luc Wolfender†, Muriel Cuendet†, and Philippe Christen*† J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b00896

|
Scooped by
NatProdChem
|
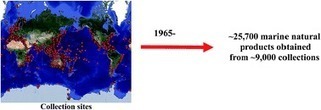
Covering: 2014. Previous review: Nat. Prod. Rep., 2015, 32, 116–211 This review covers the literature published in 2014 for marine natural products (MNPs), with 1116 citations (753 for the period January to December 2014) referring to compounds isolated from marine microorganisms and phytoplankton, green, brown and red algae, sponges, cnidarians, bryozoans, molluscs, tunicates, echinoderms, mangroves and other intertidal plants and microorganisms. The emphasis is on new compounds (1378 in 456 papers for 2014), together with the relevant biological activities, source organisms and country of origin. Reviews, biosynthetic studies, first syntheses, and syntheses that lead to the revision of structures or stereochemistries, have been included.
Nat. Prod. Rep., 2016, Advance Article
DOI: 10.1039/C5NP00156K

|
Scooped by
NatProdChem
|
A simple, robust, and inexpensive apparatus for cleaning several NMR tubes is easily fit up and used from readily available glassware, including a beaker, a desiccator, and a rotary evaporator vacuum pump.
Organic Process Research & Development (ACS Publications) Institut de Chimie des Substances Naturelles - CNRS-ICSN UPR 2301, Université Paris-Sud, 1 avenue de la Terrasse, 91198 Gif-sur-Yvette Cedex, France Org. Process Res. Dev., Article ASAP DOI: 10.1021/acs.oprd.6b00001 Publication Date (Web): January 6, 2016 Copyright © 2016 American Chemical Society

|
Scooped by
NatProdChem
|
Four limonoids, cipacinoids A–D (1–4), with spirocyclic skeletons were isolated from Cipadessa cinerascens. It is particularly notable that compounds 1–3 had a 17S-configuration for the first time in the limonoid family. Their structures with absolute configurations were assigned by spectroscopic data, X-ray crystallography, and CD analysis. Compound 1 showed moderate protein tyrosine phosphatase 1B (PTP1B) inhibition.
|

|
Scooped by
NatProdChem
|
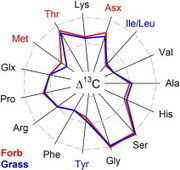
Abstract The natural carbon isotope composition of individual amino acids from plant leaf proteins has been measured to establish potential sources of variability.

|
Scooped by
NatProdChem
|
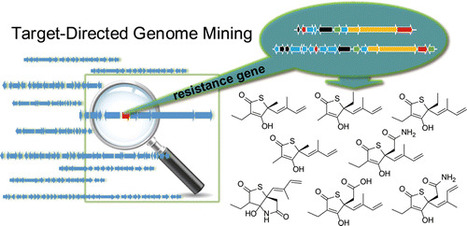
Recent genome sequencing efforts have led to the rapid accumulation of uncharacterized or “orphaned” secondary metabolic biosynthesis gene clusters (BGCs) in public databases. This increase in DNA-sequenced big data has given rise to significant challenges in the applied field of natural product genome mining, including (i) how to prioritize the characterization of orphan BGCs and (ii) how to rapidly connect genes to biosynthesized small molecules. Here, we show that by correlating putative antibiotic resistance genes that encode target-modified proteins with orphan BGCs, we predict the biological function of pathway specific small molecules before they have been revealed in a process we call target-directed genome mining. By querying the pan-genome of 86 Salinispora bacterial genomes for duplicated house-keeping genes colocalized with natural product BGCs, we prioritized an orphan polyketide synthase-nonribosomal peptide synthetase hybrid BGC (tlm) with a putative fatty acid synthase resistance gene. We employed a new synthetic double-stranded DNA-mediated cloning strategy based on transformation-associated recombination to efficiently capture tlm and the related ttm BGCs directly from genomic DNA and to heterologously express them in Streptomyces hosts. We show the production of a group of unusual thiotetronic acid natural products, including the well-known fatty acid synthase inhibitor thiolactomycin that was first described over 30 years ago, yet never at the genetic level in regards to biosynthesis and autoresistance. This finding not only validates the target-directed genome mining strategy for the discovery of antibiotic producing gene clusters without a priori knowledge of the molecule synthesized but also paves the way for the investigation of novel enzymology involved in thiotetronic acid natural product biosynthesis. † Scripps Institution of Oceanography, University of California, San Diego, La Jolla, California, United States ‡ Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, California, United States § Centro de Genómica y Bioinformática, Facultad de Ciencias, Universidad Mayor, Campus Huechuraba, Camino a la Pirámide 5750, Santiago, Chile ACS Chem. Biol., 2015, 10 (12), pp 2841–2849 DOI: 10.1021/acschembio.5b00658 Publication Date (Web): October 12, 2015

|
Scooped by
NatProdChem
|
Dereplication represents a key step for rapidly identifying known secondary metabolites in complex biological matrices. In this context, liquid-chromatography coupled to high resolution mass spectrometry (LC-HRMS) is increasingly used and, via untargeted data-dependent MS/MS experiments, massive amounts of detailed information on the chemical composition of crude extracts can be generated. An efficient exploitation of such data sets requires automated data treatment and access to dedicated fragmentation databases. Various novel bioinformatics approaches such as molecular networking (MN) and in-silico fragmentation tools have emerged recently and provide new perspective for early metabolite identification in natural products (NPs) research. Here we propose an innovative dereplication strategy based on the combination of MN with an extensive in-silico MS/MS fragmentation database of NPs. Using two case studies, we demonstrate that this combined approach offers a powerful tool to navigate through the chemistry of complex NPs extracts, dereplicate metabolites, and annotate analogues of database entries. Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.5b04804 Publication Date (Web): February 16, 2016

|
Scooped by
NatProdChem
|
Au cours de ces travaux, il a notamment isolé, à partir de 125 kg de foies et viscères de murènes, 350 µg de ciguatoxine (toxine de type polyéther d’environ 1200 de poids moléculaire responsable de l’intoxication ciguatérique ou "ciguatera" causant des troubles digestifs, neurologiques et cardio-vasculaires pouvant être graves). La ciguatoxine a été obtenue avec une pureté de l’ordre de 95%, ce qui a permis au Prof. T. Yasumoto d’en déterminer la structure et sa configuration (1989 et 1990).

|
Scooped by
NatProdChem
|
Marine and terrestrial organisms yield a remarkable chemical diversity and are important sources for discovery of new chemical products. In order to maximize the bioprospecting efficiency of natural products (NP), taxonomy, geography and biodiversity are starting to be used to draw conclusions on which taxonomic groups and/or regions may be of interest for future research. However, accurate taxonomic information and sampling location of source organisms have often been overlooked. Although these issues were already reported a few decades ago and improvements have been made, such outstanding problems are still recurrent in recent peer-reviewed literature. Here, we focus on the importance of taxonomic and geographic identification of source material and illustrate how taxonomic and geographic data of source organisms continues to be poorly handled. It is our opinion that this issue needs to be discussed within the NP community with the ultimate goal of improving publication standards and guaranteeing the scientific principle of research reproducibility. Moreover, by doing so, it will be possible to take advantage of information available in the literature to develop cross-disciplinary meta-analyses that may help to advance the state of the art of NP research and future bioprospecting endeavours.
Nat. Prod. Rep., 2016, Advance Article
DOI: 10.1039/C5NP00130G

|
Scooped by
NatProdChem
|
3-Acylated tetramic and tetronic acids are characterized by a low pKa and are likely to be deprotonated under physiological conditions. In addition, their structure makes them excellent chelators of metallic cations. We will discuss the significance of these chemical properties with regard to the biological properties and mechanisms of action of these compounds, highlighting the importance of considering them as salts or chelates for biological purposes, rather than acids.
Nat. Prod. Rep., 2016, Advance Article
DOI: 10.1039/C5NP00144G

|
Scooped by
NatProdChem
|
A chemical investigation of the tropical sponge Agelas sceptrum from Plana Cays (Bahamas) led to the isolation of two hybrid pyrrole–imidazole alkaloids (PIAs), 15′-oxoadenosceptrin (1) and decarboxyagelamadin C (2). Herein, we report their challenging structure elucidation established by NMR and ECD spectroscopy. 15′-Oxoadenosceptrin (1) shows sceptrin merged with an adenine moiety, not yet encountered in the PIA family, whereas decarboxyagelamadin C (2) is a close derivative of agelamadins C to E recently isolated from an Agelas sp. from Okinawa
J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b00265

|
Scooped by
NatProdChem
|
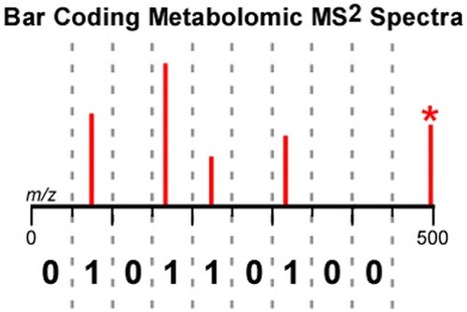
Metabolite identifications are most frequently achieved in untargeted metabolomics by matching precursor mass and full, high-resolution MS2 spectra to metabolite databases and standards. Here we considered an alternative approach for establishing metabolite identifications that does not rely on full, high-resolution MS2 spectra. First, we select mass-to-charge regions containing the most informative metabolite fragments and designate them as bins. We then translate each metabolite fragmentation pattern into a binary code by assigning 1’s to bins containing fragments and 0’s to bins without fragments. With 20 bins, this binary-code system is capable of distinguishing 96% of the compounds in the METLIN MS2 library. A major advantage of the approach is that it extends untargeted metabolomics to low-resolution triple quadrupole (QqQ) instruments, which are typically less expensive and more robust than other types of mass spectrometers. We demonstrate a method of acquiring MS2 data in which the third quadrupole of a QqQ instrument cycles over 20 wide isolation windows (coinciding with the location and width of our bins) for each precursor mass selected by the first quadrupole. Operating the QqQ instrument in this mode yields diagnostic bar codes for each precursor mass that can be matched to the bar codes of metabolite standards. Furthermore, our data suggest that using low-resolution bar codes enables QqQ instruments to make MS2-based identifications in untargeted metabolomics with a specificity and sensitivity that is competitive to high-resolution time-of-flight technologies. † Department of Chemistry, Washington University in St. Louis, St. Louis, Missouri 63130, United States ‡ Department of Medicine, Washington University School of Medicine, St. Louis, Missouri 63110, United States § Department of Genetics, Washington University School of Medicine, St. Louis, Missouri 63110, United States Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.5b04925

|
Scooped by
NatProdChem
|
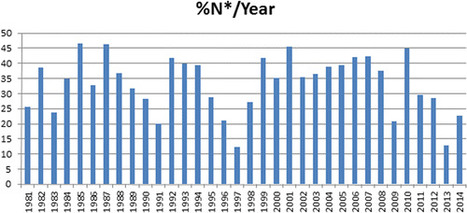
This contribution is a completely updated and expanded version of the four prior analogous reviews that were published in this journal in 1997, 2003, 2007, and 2012. In the case of all approved therapeutic agents, the time frame has been extended to cover the 34 years from January 1, 1981, to December 31, 2014, for all diseases worldwide, and from 1950 (earliest so far identified) to December 2014 for all approved antitumor drugs worldwide. As mentioned in the 2012 review, we have continued to utilize our secondary subdivision of a “natural product mimic”, or “NM”, to join the original primary divisions and the designation “natural product botanical”, or “NB”, to cover those botanical “defined mixtures” now recognized as drug entities by the U.S. FDA (and similar organizations). From the data presented in this review, the utilization of natural products and/or their novel structures, in order to discover and develop the final drug entity, is still alive and well. For example, in the area of cancer, over the time frame from around the 1940s to the end of 2014, of the 175 small molecules approved, 131, or 75%, are other than “S” (synthetic), with 85, or 49%, actually being either natural products or directly derived therefrom. In other areas, the influence of natural product structures is quite marked, with, as expected from prior information, the anti-infective area being dependent on natural products and their structures. We wish to draw the attention of readers to the rapidly evolving recognition that a significant number of natural product drugs/leads are actually produced by microbes and/or microbial interactions with the “host from whence it was isolated”, and therefore it is considered that this area of natural product research should be expanded significantly.
† NIH Special Volunteer, Wayne, Pennsylvania 19087, United States ‡ NIH Special Volunteer, Bethesda, Maryland 20814, United States J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b01055

|
Scooped by
NatProdChem
|
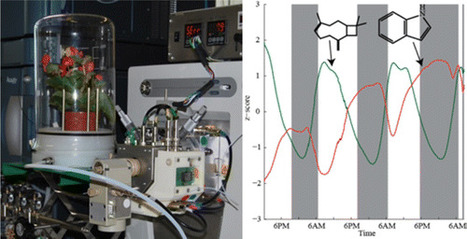
We have deployed an efficient secondary electrospray ionization source coupled to an Orbitrap mass analyzer (SESI-MS) to investigate the emissions of a Begonia semperflorens. We document how hundreds of species can be tracked with an unparalleled time resolution of 2 min during day–night cycles. To further illustrate the capabilities of this system for volatile organic compounds (VOCs) analysis, we subjected the plant to mechanical damage and monitored its response. As a result, ∼1200 VOCs were monitored displaying different kinetics. To validate the soundness of our in vivo measurements, we fully characterized some key compounds via tandem mass spectrometry (MS/MS) and confirmed their expected behavior based on prior gas chromatography/mass spectrometry (GC/MS) studies. For example, β-caryophyllene, which is directly related to photosynthesis, was found to show a periodic day–night pattern with highest concentrations during the day. We conclude that the capability of SESI-MS to capture highly dynamic VOC emissions and wide analyte coverage makes it an attractive tool to complement GC/MS in plant studies.
Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.5b04452

|
Scooped by
NatProdChem
|
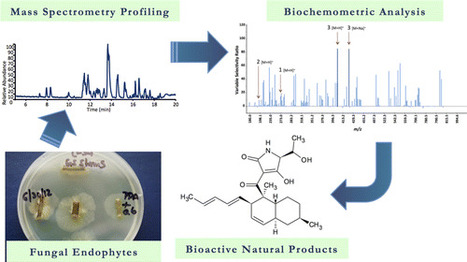
A central challenge of natural products research is assigning bioactive compounds from complex mixtures. The gold standard approach to address this challenge, bioassay-guided fractionation, is often biased toward abundant, rather than bioactive, mixture components. This study evaluated the combination of bioassay-guided fractionation with untargeted metabolite profiling to improve active component identification early in the fractionation process. Key to this methodology was statistical modeling of the integrated biological and chemical data sets (biochemometric analysis). Three data analysis approaches for biochemometric analysis were compared, namely, partial least-squares loading vectors, S-plots, and the selectivity ratio. Extracts from the endophytic fungi Alternaria sp. and Pyrenochaeta sp. with antimicrobial activity against Staphylococcus aureus served as test cases. Biochemometric analysis incorporating the selectivity ratio performed best in identifying bioactive ions from these extracts early in the fractionation process, yielding altersetin (3, MIC 0.23 μg/mL) and macrosphelide A (4, MIC 75 μg/mL) as antibacterial constituents from Alternaria sp. and Pyrenochaeta sp., respectively. This study demonstrates the potential of biochemometrics coupled with bioassay-guided fractionation to identify bioactive mixture components. A benefit of this approach is the ability to integrate multiple stages of fractionation and bioassay data into a single analysis
† Department of Chemistry & Biochemistry, University of North Carolina Greensboro, Greensboro, North Carolina 27402, United States ‡ Department of Chemistry, University of Bergen, Bergen 5020,Norway J. Nat. Prod., Article ASAP

|
Scooped by
NatProdChem
|
- First marketing authorization of Dengvaxia® is a historic milestone paving the way
to significantly impact dengue burden in endemic countries -
|



 Your new post is loading...
Your new post is loading...



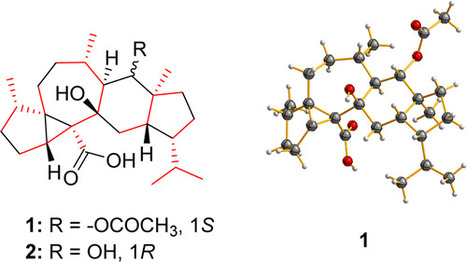












Very interseting article by Cichewicz's team, were the role of a fungal secondary metabolite is explored in detail.
Very interseting article by Cichewicz's team, were the role of a fungal secondary metabolite is explored in detail.