Your new post is loading...
 Your new post is loading...
Author: Daniel J. Cosgrove.
Annual Review of Plant Cell and Development (2024)
Abstract: "Expansins comprise an ancient group of cell wall proteins ubiquitous in land plants and their algal ancestors. During cell growth, they facilitate passive yielding of the wall's cellulose networks to turgor-generated tensile stresses, without evidence of enzymatic activity. Expansins are also implicated in fruit softening and other developmental processes and in adaptive responses to environmental stresses and pathogens. The major expansin families in plants include α-expansins (EXPAs), which act on cellulose-cellulose junctions, and β-expansins, which can act on xylans. EXPAs mediate acid growth, which contributes to wall enlargement by auxin and other growth agents. The genomes of diverse microbes, including many plant pathogens, also encode expansins designated expansin-like X. Expansins are proposed to disrupt noncovalent bonding between laterally aligned polysaccharides (notably cellulose), facilitating wall loosening for a variety of biological roles."
Authors: Jun-Li Wang, Ming Wang, Li Zhang, You-Xia Li, Jing-Jing Li, Yu-Yang Li, Zuo-Xian Pu, Dan-Yang Li, Xing-Nan Liu, Wang Guo, Dong-Wei Di, Xiao-Feng Li, Guang-Qin Guo and Lei Wu.
PNAS (2024)
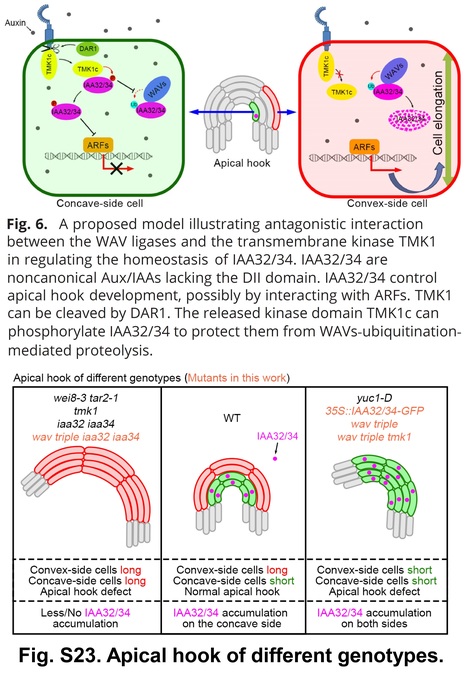
Significance: The apical hook protects cotyledons and shoot apical meristem from injuries during seedling emergence from the soil. Auxin plays a key regulatory role in hook formation. Recently, a noncanonical auxin signaling pathway was discovered during apical hook development. Asymmetrical accumulation of auxin in the hook on its concave side triggers cleavage of the C-terminal of TMK1, which then travels to the nucleus and stabilizes two noncanonical Aux/IAAs (IAA32/34) by phosphorylation. Phosphorylated IAA32/34 inhibit elongation of cells on the concave side, resulting in hook formation. This work shows that IAA32/34 degradation is initiated through ubiquitination by WAV E3 ligases, which is inhibited by TMK1-mediated phosphorylation. Our studies identified components critical for IAA32/34 turnover in this TMK1-mediated auxin signaling pathway.
Abstract: "Auxin regulates plant growth and development through downstream signaling pathways, including the best-known SCFTIR1/AFB-Aux/IAA-ARF pathway and several other less characterized “noncanonical” pathways. Recently, one SCFTIR1/AFB-independent noncanonical pathway, mediated by Transmembrane Kinase 1 (TMK1), was discovered through the analyses of its functions in Arabidopsis apical hook development. Asymmetric accumulation of auxin on the concave side of the apical hook triggers DAR1-catalyzed release of the C-terminal of TMK1, which migrates into the nucleus, where it phosphorylates and stabilizes IAA32/34 to inhibit cell elongation, which is essential for full apical hook formation. However, the molecular factors mediating IAA32/34 degradation have not been identified. Here, we show that proteins in the CYTOKININ INDUCED ROOT WAVING 1 (CKRW1)/WAVY GROWTH 3 (WAV3) subfamily act as E3 ubiquitin ligases to target IAA32/34 for ubiquitination and degradation, which is inhibited by TMK1c-mediated phosphorylation. This antagonistic interaction between TMK1c and CKRW1/WAV3 subfamily E3 ubiquitin ligases regulates IAA32/34 levels to control differential cell elongation along opposite sides of the apical hook."
Authors: Yuyuan Ma, Yu Wang, Zhiqin Zhou, Runqin Zhang, Yiru Xie, Yihan Zhang, Yongming Bo, Xiaolong Lyu, Jinghua Yang, Mingfang Zhang and Zhongyuan Hu.
Theoretical and Applied Genetics (2024)
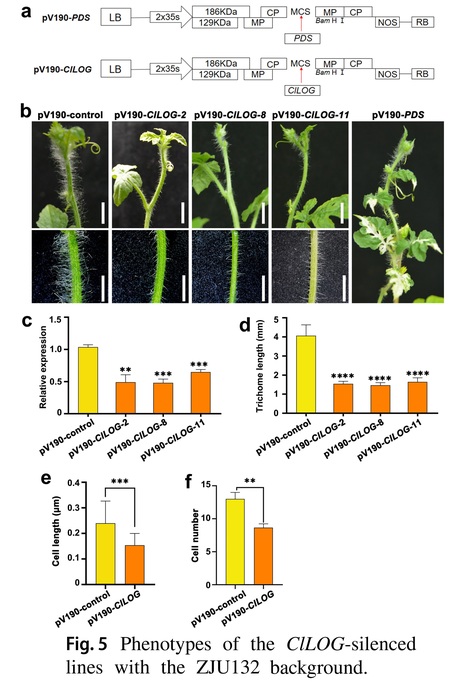
Key message: The ClLOG gene encoding a cytokinin riboside 5ʹ-monophosphate phosphoribohydrolase determines trichome length in watermelon, which is associated with its promoter variations.
Abstract: "Trichomes, which are differentiated from epidermal cells, are special accessory structures that cover the above-ground organs of plants and possibly contribute to biotic and abiotic stress resistance. Here, a bulked segregant analysis (BSA) of an F2 population with significant variations in trichome length was undertaken. A 1.84-Mb candidate region on chromosome 10 was associated with trichome length. Resequencing and fine-mapping analyses indicated that a 12-kb structural variation in the promoter of Cla97C10G203450 (ClLOG) led to a significant expression difference in this gene in watermelon lines with different trichome lengths. In addition, a virus-induced gene silencing analysis confirmed that ClLOG positively regulated trichome elongation. These findings provide new information and identify a potential target gene for controlling multicellular trichome elongation in watermelon."
Authors: Zhuang Li, Qichao Tu, Xiangguang Lyu, Qican Cheng, Ronghuan Ji, Chao Qin, Jun Liu, Bin Liu, Hongyu Li and Tao Zhao.
The Crop Journal (2024)
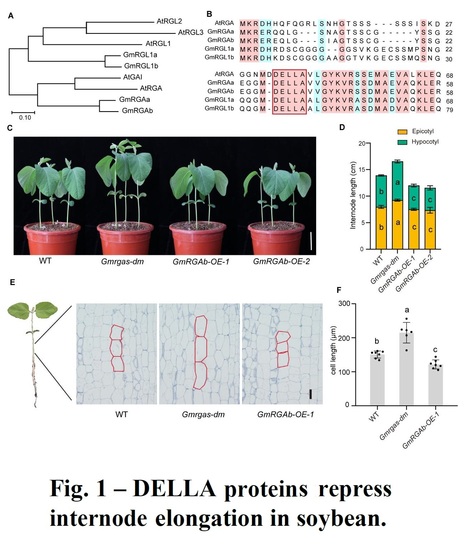
Abstract: "Plant height influences plant architecture, lodging resistance, and yield performance. It is modulated by gibberellic acid (GA) metabolism and signaling. DELLA proteins, acting as central repressors of GA signaling, integrate various environmental and hormonal signals to regulate plant growth and development in Arabidopsis. We examined the role of two DELLA proteins, GmRGAa and GmRGAb, in soybean plant height control. Knockout of these proteins led to longer internodes and increased plant height, primarily by increasing cell elongation. GmRGAs functioned under different light conditions, including red, blue, and far-red light, to repress plant height. Interaction studies revealed that GmRGAs interacted with the blue light receptor GmCRY1b. Consistent with this, GmCRY1b partially regulated plant height via GmRGAs. Additionally, DELLA proteins were found to stabilize the protein GmSTF1/2, a key positive regulator of photomorphogenesis. This stabilization led to increased transcription of GmGA2ox-7b and subsequent reduction in plant height. This study enhances our understanding of DELLA-mediated plant height control, offering Gmrgaab mutants for soybean structure and yield optimization.
Authors: Sonali Roy, Ivone Torres-Jerez, Shulan Zhang, Wei Liu, Katharina Schiessl, Divya Jain, Clarissa Boschiero, Hee-Kyung Lee, Nicholas Krom, Patrick X. Zhao, Jeremy D. Murray, Giles E. D. Oldroyd, Wolf-Rüdiger Scheible and Michael Udvardi.
The Plant Journal (2024)
Significance Statement: Nodule positioning is an understudied trait, yet it determines the length of the root that can support nodule formation and consequently the total number of functional nodules formed. We identify genetic factors called GOLVEN peptides that alter nodule and lateral root positioning on the primary root along with several other traits including nodule organ initiation and root architecture.
Abstract: "The conservation of GOLVEN (GLV)/ROOT MERISTEM GROWTH FACTOR (RGF) peptide encoding genes across plant genomes capable of forming roots or root-like structures underscores their potential significance in the terrestrial adaptation of plants. This study investigates the function and role of GOLVEN peptide-coding genes in Medicago truncatula. Five out of fifteen GLV/RGF genes were notably upregulated during nodule organogenesis and were differentially responsive to nitrogen deficiency and auxin treatment. Specifically, the expression of MtGLV9 and MtGLV10 at nodule initiation sites was contingent upon the NODULE INCEPTION transcription factor. Overexpression of these five nodule-induced GLV genes in hairy roots of M. truncatula and application of their synthetic peptide analogues led to a decrease in nodule count by 25–50%. Uniquely, the GOLVEN10 peptide altered the positioning of the first formed lateral root and nodule on the primary root axis, an observation we term ‘noduletaxis’; this decreased the length of the lateral organ formation zone on roots. Histological section of roots treated with synthetic GOLVEN10 peptide revealed an increased cell number within the root cortical cell layers without a corresponding increase in cell length, leading to an elongation of the root likely introducing a spatiotemporal delay in organ formation. At the transcription level, the GOLVEN10 peptide suppressed expression of microtubule-related genes and exerted its effects by changing expression of a large subset of Auxin responsive genes. These findings advance our understanding of the molecular mechanisms by which GOLVEN peptides modulate root morphology, nodule ontogeny, and interactions with key transcriptional pathways."
Authors: Peng He, Liping Zhu, Xin Zhou, Xuan Fu, Yu Zhang, Peng Zhao, Bin Jiang, Huiqin Wang and Guanghui Xiao.
Developmental Cell (2024)
Editor's view: He et al. identified two signaling cascades, GhSLR1-GhZFP8-GhSDCP1- GhPIF3 and GhSLR1-GhBLH1-GhKCS12, which translate the gibberellins signal to regulate cotton fiber cell elongation. This finding can be potentially used in the field to improve cotton fiber quality.
Highlight: • Gibberellins promotes fiber cell elongation by degrading GhSLR1 • GhSLR1 interacts with two transcription factors, GhZFP8 and GhBLH1 • GhBLH1 activates the transcription of GhKCS12 and enhances VLCFA biosynthesis • GhZFP8 activates the expression of GhPIF3 by upregulating GhSDCP1 in cotton
Abstract: "The agricultural green revolution spectacularly enhanced crop yield through modification of gibberellin (GA) signaling. However, in cotton, the GA signaling cascades remain elusive, limiting our potential to cultivate new cotton varieties and improve yield and quality. Here, we identified that GA prominently stimulated fiber elongation through the degradation of DELLA protein GhSLR1, thereby disabling GhSLR1’s physical interaction with two transcription factors, GhZFP8 and GhBLH1. Subsequently, the resultant free GhBLH1 binds to GhKCS12 promoter and activates its expression to enhance VLCFAs biosynthesis. With a similar mechanism, the free GhZFP8 binds to GhSDCP1 promoter and activates its expression. As a result, GhSDCP1 upregulates the expression of GhPIF3 gene associated with plant cell elongation. Ultimately, the two parallel signaling cascades synergistically promote cotton fiber elongation. Our findings outline the mechanistic framework that translates the GA signal into fiber cell elongation, thereby offering a roadmap to improve cotton fiber quality and yield."
Authors: Esther Cañibano, Daniela Soto-Gomez, Juan Carlos Oliveros, Clara Bourbousse and Sandra Fonseca.
bioRxiv (2024)
Abstract: "Driven by cell elongation, hypocotyl growth is tightly controlled by light and responds to external stimuli and endogenous hormonal pathways. Hypocotyls are known to be responsive to the stress signalling hormone abscisic acid (ABA) which effectively inhibits cell elongation, but how this regulation is connected to light responses and other endogenous hormonal pathways has been a subject of limited studies. Here, we show that whereas hypocotyl elongation is sensitive to ABA in light-grown seedlings, the hypocotyl of dark-grown etiolated seedlings is ABA-insensitive. In the dark, hypocotyl sensitivity to ABA is restored in the constitutive photomorphogenic pifq and cop1-4 mutants, suggesting that an active light signalling pathway is necessary for hypocotyl responsiveness to ABA. However, etiolated hypocotyls retain ABA responsiveness, as could be detected by the induction of ABI1 and RD29B transcripts in response to exogenous ABA, suggesting that inhibition of hypocotyl elongation mediated by ABA does not follows the canonical ABA signalling dependent on transcription. Here, using RNA-seq analysis we identified a number of ABA differentially expressed genes (DEGs) that correlate with ABA inhibition of hypocotyl elongation, specifically in dark-grown pifq or light-grown WT plants, and whose expression remains unchanged by ABA treatment in dark-grown WT plants. Among these DEGs we identified a number of genes playing a role in cell elongation directly at the level of the plasma membrane, as SAURs, ion transporters, auxin flux regulators, channels, and cell wall modification enzymes. The use of the auxin transport inhibitor, NPA, revealed that in the light auxin transport impairment renders hypocotyls insensitive to ABA in WT and pifq plants. Thus, in the light, hypocotyl responsiveness to ABA is dependent on auxin transport and independent of PIFs. In the dark, PIFs render hypocotyls insensitive to ABA, perhaps by regulating the expression of a number of ABA DEGs, a mechanism that could allow plants to prioritize the elongation towards light, avoiding to slow-down soil emergence that could be induced by ABA signalling in case of sudden reduction of soil moisture."
Authors: Xueya Zhao, Kunpeng Zhang, Huidong Zhang, Mengxi Bi, Yi He, Yiqing Cui, Changhua Tan, Jian Ma and Mingfang Qi.
Frontiers in Plant Science (2023)
Abstract: "Plant height is an important agronomic trait. Dwarf varieties present several advantages, such as lodging resistance, increased yield, and suitability for mechanized harvesting, which are crucial for crop improvement. However, limited research is available on dwarf tomato varieties suitable for production. In this study, we report a novel short internode mutant named “short internode and pedicel (sip)” in tomato, which exhibits marked internode and pedicel shortening due to suppressed cell elongation. This mutant plant has a compact plant structure and compact inflorescence, and has been demonstrated to produce more fruits, resulting in a higher harvest index. Genetic analysis revealed that this phenotype is controlled by a single recessive gene, SlSIP. BSA analysis and KASP genotyping indicated that ERECTA (ER) is the possible candidate gene for SlSIP, which encodes a leucine-rich receptor-like kinase. Additionally, we obtained an ER functional loss mutant using the CRISPR/Cas9 gene-editing technology. The 401st base A of ER is substituted with T in sip, resulting in a change in the 134th amino acid from asparagine (N) to isoleucine (I). Molecular dynamics(MD) simulations showed that this mutation site is located in the extracellular LRR domain and alters nearby ionic bonds, leading to a change in the spatial structure of this site. Transcriptome analysis indicated that the genes that were differentially expressed between sip and wild-type (WT) plants were enriched in the gibberellin metabolic pathway. We found that GA3 and GA4 decreased in the sip mutant, and exogenous GA3 restored the sip to the height of the WT plant. These findings reveal that SlSIP in tomatoes regulates stem elongation by regulating gibberellin metabolism. These results provide new insights into the mechanisms of tomato dwarfing and germplasm resources for breeding dwarfing tomatoes."
Authors: Keming Luo, Shuai Liu, Xiaokang Fu, Xuelian Du, Jian Hu, Lianjia Luo, Changjian Fa, Rongling Wu, Laigeng Li and Changzheng Xu.
Research Square (2023)
Abstract: "Auxin, as a vital phytohormone, is enriched in the vascular cambium, playing a crucial role in regulating wood formation in trees. Despite its significance, the molecular mechanisms underlying the influence of auxin on wood development remain elusive. In this study, we report a transcription factor, PLETHORA 5 (PLT5), whose expression was specifically activated by auxin signalling in the vascular cambium. PLT5 was found to regulate cell expansion and lignification of fibres in poplar. Genetic experiments confirmed the noncell-autonomous regulation of auxin signalling from the vascular cambium and revealed the necessity of PLT5 protein mobility to mediate this process. Remarkably, PLT5 proteins specifically inhibit the initiation of fibre cell wall thickening by directly repressing SND1 genes. This study unveils a sophisticated model wherein the auxin-PLT5 signalling cascade intricately regulates wood fibre development in poplar by fine-tuning the thickening of fibre cell walls."
Authors: Xueliang Zhang, Zhikang Shen, Xiaohu Sun, Min Chen and Naichao Zhang.
Functional Plant Biology (2023)
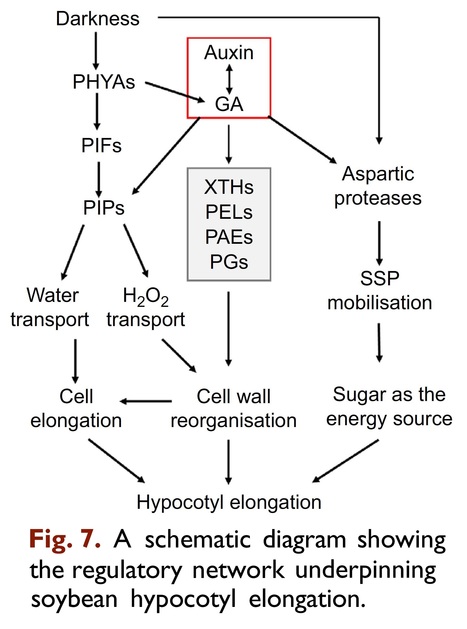
Abstract: "Hypocotyl elongation directly affects the seedling establishment and soil-breaking after germination. In soybean (Glycine max), the molecular mechanisms regulating hypocotyl development remain largely elusive. To decipher the regulatory landscape, we conducted proteome and transcriptome analysis of soybean hypocotyl samples at different development stages. Our results showed that during hypocotyl development, many proteins were with extreme high translation efficiency (TE) and may act as regulators. These potential regulators include multiple peroxidases and cell wall reorganisation related enzymes. Peroxidases may produce ROS including H2O2. Interestingly, exogenous H2O2 application promoted hypocotyl elongation, supporting peroxidases as regulators of hypocotyl development. However, a vast variety of proteins were shown to be with dramatically changed TE during hypocotyl development, including multiple phytochromes, plasma membrane intrinsic proteins (PIPs) and aspartic proteases. Their potential roles in hypocotyl development were confirmed by that ectopic expression of GmPHYA1 and GmPIP1-6 in Arabidopsis thaliana affected hypocotyl elongation. In addition, the promoters of these potential regulatory genes contain multiple light/gibberellin/auxin responsive elements, while the expression of some members in hypocotyls was significantly regulated by light and exogenous auxin/gibberellin. Overall, our results revealed multiple novel regulatory factors of soybean hypocotyl elongation. Further research on these regulators may lead to new approvals to improve soybean hypocotyl traits."
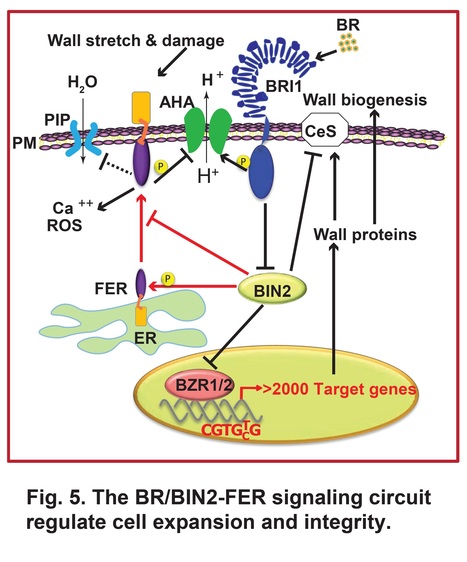
Authors: Ajeet Chaudhary, Yu-Chun Hsiao, Fang-Ling Jessica Yeh, Hen-Ming Wu, Alice Y. Cheung, Shou-Ling Xu and Zhi-Yong Wang.
bioRxiv (2023)
One-sentence summary: Brassinosteroid recruits a cell wall integrity monitor to prevent growth-induced cell wall damage.
Abstract: "Plant cell expansion is driven by turgor pressure and regulated by hormones. How plant cells avoid cell wall rupture during hormone-induced cell expansion remains a mystery. Here we show that brassinosteroid (BR), while stimulating cell elongation, promotes the plasma membrane (PM) accumulation of the receptor kinase FERONIA (FER), which monitors cell wall damage and in turn attenuates BR-induced cell elongation to prevent cell rupture. The GSK3-like kinase BIN2 phosphorylates FER, resulting in reduced FER accumulation and translocation from endoplasmic reticulum to PM. By inactivating BIN2, BR signaling promotes dephosphorylation and increases PM accumulation of FER, thereby enhancing the surveillance of cell wall integrity. Our study reveals a vital signaling circuit that coordinates hormone signaling with mechanical sensing to prevent cell bursting during hormone-induced cell expansion."
Authors: Jing Nie, Yu Jiang, Lijun Lv, Yuzi Shi, Peiyu Chen, Qian Zhang and Xiaolei Sui.
Horticultural Plant Journal (2023)
Abstract: "High temperature-induced hypocotyl elongation is a typical thermomorphogenesis trait that may significantly affect early seedling growth and subsequent crop yield. The ambient temperature and endogenous auxin are two critical factors that regulate hypocotyl growth. However, the mechanism of temperature and auxin integration in horticultural plants remains poorly understood. In this study, the roles of the basic helix-loop-helix transcription factor CsPIF4 in regulating auxin biosynthesis genes and the auxin content in the hypocotyl of cucumber (Cucumis sativus L.) seedlings under high temperature were investigated. qRT-PCR and in situ hybridization analysis revealed that expression of CsPIF4 was enhanced in the epidermis and vascular bundles in the hypocotyl of cucumber seedlings in response to high temperature. qRT-PCR and HPLC analysis showed that CsPIF4 positively regulated transcription of the auxin biosynthesis gene CsYUC8 and the auxin content in the hypocotyl under high temperature (35 °C). The CRISPR/Cas9-mediated knockout of CsPIF4 resulted in a shorter hypocotyl compared with that of the wild type, together with decreased expression of CsYUC8 and lower auxin content in response to high temperature. Furthermore, biochemical assays showed that CsPIF4 could bind directly to the G-box motif of the CsYUC8 promoter and thereby activate CsYUC8 expression. These findings provide insight into the molecular mechanism of high temperature-mediated hypocotyl elongation in cucumber."
Authors: Wenyi Wang, Vanika Garg, Rajeev K. Varshney and Hao Liu.
Trends in Plant Science (2023).
Abstract: Phytohormone signaling regulates plant growth and development. Single cell RNA sequencing (scRNA-seq) provides unprecedented opportunities to decipher hormone-mediated spatiotemporal gene regulatory networks. In a recent study, Nolan et al. used time-series scRNA-seq to identify the cortex as a key site for brassinosteroid (BR)-mediated gene expression and revealed a signaling network during cell phase transition."
|
Authors: Wenli Hu, Rong Wang, Xiaohua Hao, Shaozhuang Li, Xinjie Zhao, Zijing Xie, Sha Wu, Liqun Huang, Ying Tan, Lianfu Tian and Dongping Li.
The Plant Journal (2024)
Significant Statement: The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight.
Abstract: "A fundamental question in developmental biology is how to regulate grain size to improve crop yields. Despite this, little is still known about the genetics and molecular mechanisms regulating grain size in crops. Here, we provide evidence that a putative protein kinase-like (OsLCD3) interacts with the S-adenosyl-L-methionine synthetase 1 (OsSAMS1) and determines the size and weight of grains. OsLCD3 mutation (lcd3) significantly increased grain size and weight by promoting cell expansion in spikelet hull, whereas its overexpression caused negative effects, suggesting that grain size was negatively regulated by OsLCD3. Importantly, lcd3 and OsSAMS1 overexpression (SAM1OE) led to large and heavy grains, with increased ethylene and decreased polyamines production. Based on genetic analyses, it appears that OsLCD3 and OsSAMS1 control rice grain size in part by ethylene/polyamine homeostasis. The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight."
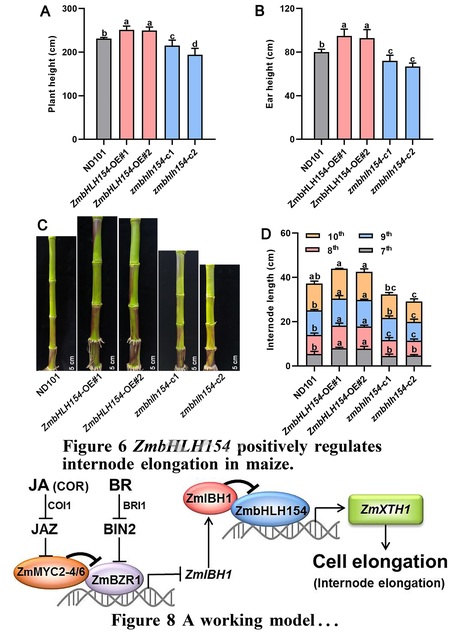
Authors: Xing Wang, Zhaobin Ren, Shipeng Xie, Zhaohu Li, Yuyi Zhou and Liusheng Duan.
Plant Physiology (2024)
One-sentence summary: A jasmonate mimic regulates a basic helix-loop-helix network by attenuating brassinosteroid signaling, which represses expression of a cell wall–related gene and inhibits internode elongation in maize.
Abstract: "Lodging restricts growth, development, and yield formation in maize (Zea mays L.). Shorter internode length is beneficial for lodging tolerance. However, although brassinosteroids (BRs) and jasmonic acid (JA) are known to antagonistically regulate internode growth, the underlying molecular mechanism is still unclear. In this study, application of the JA mimic coronatine (COR) inhibited basal internode elongation at the jointing stage and repressed expression of the cell wall-related gene XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 1 (ZmXTH1), whose overexpression in maize plants promotes internode elongation. We demonstrated that the basic helix–loop–helix (bHLH) transcription factor ZmbHLH154 binds directly to the ZmXTH1 promoter and induces its expression, whereas the bHLH transcription factor ILI1 BINDING BHLH 1 (ZmIBH1) inhibits this transcriptional activation by forming a heterodimer with ZmbHLH154. Overexpressing ZmbHLH154 led to longer internodes, whereas zmbhlh154 mutants had shorter internodes than the wild type. The core JA-dependent transcription factors ZmMYC2-4 and ZmMYC2-6 interacted with BRASSINAZOLE RESISTANT 1 (ZmBZR1), a key factor in BR signaling, and these interactions eliminated the inhibitory effect of ZmBZR1 on its downstream gene ZmIBH1. Collectively, these results reveal a signaling module in which JA regulates a bHLH network by attenuating BR signaling to inhibit ZmXTH1 expression, thereby regulating cell elongation in maize."
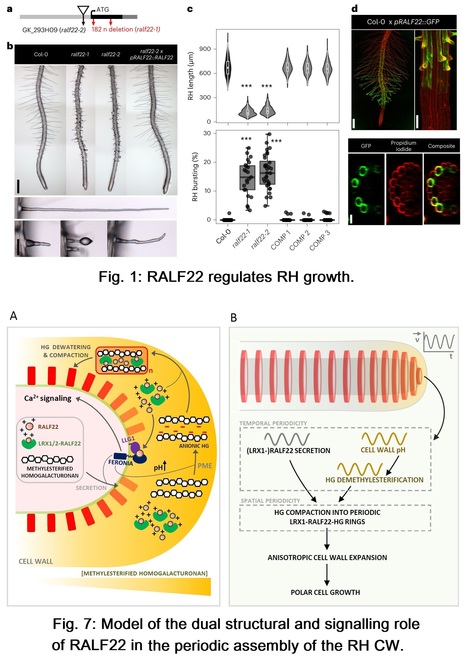
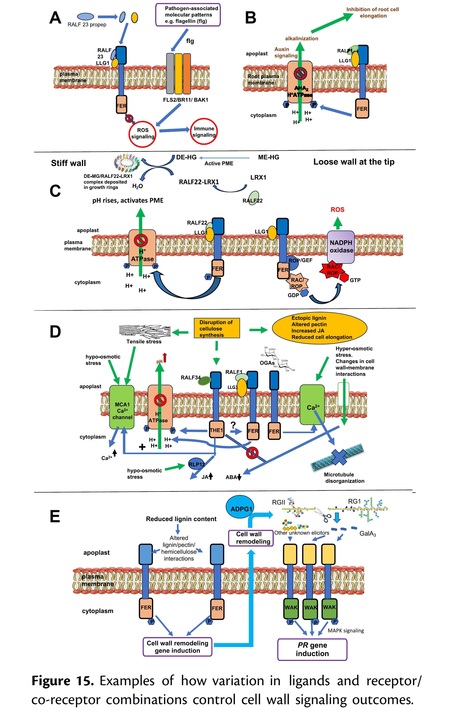
Authors: Sébastjen Schoenaers, Hyun Kyung Lee, Martine Gonneau, Elvina Faucher, Thomas Levasseur, Elodie Akary, Naomi Claeijs, Steven Moussu, Caroline Broyart, Daria Balcerowicz, Hamada AbdElgawad, Andrea Bassi, Daniel Santa Cruz Damineli, Alex Costa, José A. Feijó, Celine Moreau, Estelle Bonnin, Bernard Cathala, Julia Santiago, Herman Höfte and Kris Vissenberg.
Nature Plants (2024)
Editor´s view: The authors show that RALF22 has a dual role in cell wall assembly in root hairs: as a structural component organizing cell wall architecture and as a feedback signalling molecule that regulates this process depending on its interaction partners.
Abstract: "Pressurized cells with strong walls make up the hydrostatic skeleton of plants. Assembly and expansion of such stressed walls depend on a family of secreted RAPID ALKALINIZATION FACTOR (RALF) peptides, which bind both a membrane receptor complex and wall-localized LEUCINE-RICH REPEAT EXTENSIN (LRXs) in a mutually exclusive way. Here we show that, in root hairs, the RALF22 peptide has a dual structural and signalling role in cell expansion. Together with LRX1, it directs the compaction of charged pectin polymers at the root hair tip into periodic circumferential rings. Free RALF22 induces the formation of a complex with LORELEI-LIKE-GPI-ANCHORED PROTEIN 1 and FERONIA, triggering adaptive cellular responses. These findings show how a peptide simultaneously functions as a structural component organizing cell wall architecture and as a feedback signalling molecule that regulates this process depending on its interaction partners. This mechanism may also underlie wall assembly and expansion in other plant cell types."
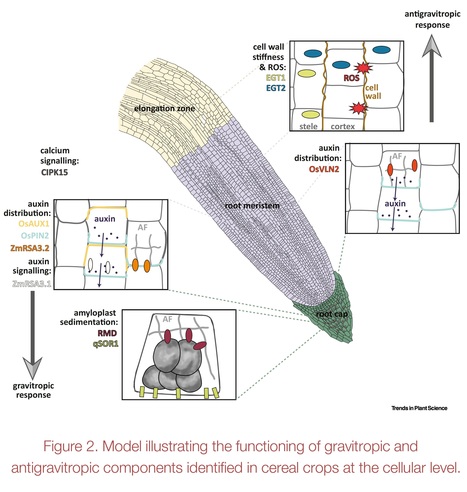
Authors: Gwendolyn K. Kirschner, Frank Hochholdinger, Silvio Salvi, Malcolm J. Bennett, Guoqiang Huang and Rahul A. Bhosale.
Trends in Plant Science (2024)
Highlights: The root angle in cereals determines soil resource capture, stress resilience, and yield, especially in suboptimal conditions. Root angle regulation involves competing gravitropic and antigravitropic offset mechanisms. Understanding the mechanisms underlying root angle regulation in cereals is important due to their complex root system made up of distinct root types, formed at different stages of development. Recent studies in cereals revealed genes regulating the root angle. However, the precise mechanisms determining and maintaining root angle in distinct root types remain unclear. Understanding the molecular mechanisms underlying root angle control is essential for incorporating the root angle trait into breeding programs.
Abstract: "The root angle plays a critical role in efficiently capturing nutrients and water from different soil layers. Steeper root angles enable access to mobile water and nitrogen from deeper soil layers, whereas shallow root angles facilitate the capture of immobile phosphorus from the topsoil. Thus, understanding the genetic regulation of the root angle is crucial for breeding crop varieties that can efficiently capture resources and enhance yield. Moreover, this understanding can contribute to developing varieties that effectively sequester carbon in deeper soil layers, supporting global carbon mitigation efforts. Here we review and consolidate significant recent discoveries regarding the molecular components controlling root angle in cereal crop species and outline the remaining research gaps in this field."
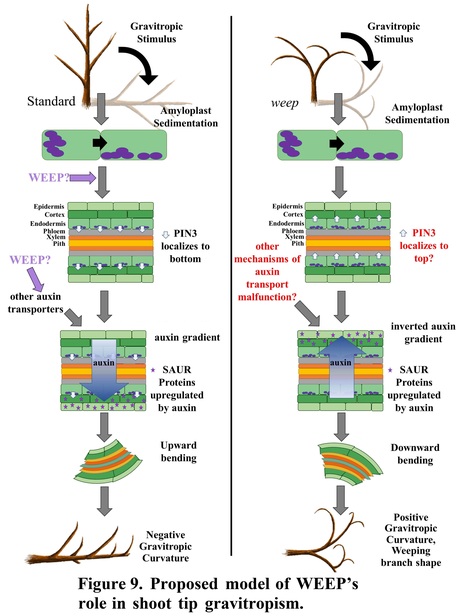
Authors: Andrea R. Kohler, Andrew Scheil, Joseph L. Hill, Jr., Jeffrey R. Allen, Jameel M. Al-Haddad, Charity Z. Goeckeritz, Lucia C. Strader, Frank W. Telewski and Courtney A. Hollender.
Plant Physiology (2024)
One-sentence summary: Polar auxin transport associated with gravitropism and lateral shoot and root orientation requires the highly conserved WEEP protein in peach.
Abstract: "Trees with weeping shoot architectures are valued for their beauty and are a resource for understanding how plants regulate posture control. The peach (Prunus persica) weeping phenotype, which has elliptical downward arching branches, is caused by a homozygous mutation in the WEEP gene. Little is known about the function of WEEP despite its high conservation throughout Plantae. Here, we present the results of anatomical, biochemical, biomechanical, physiological, and molecular experiments that provide insight into WEEP function. Our data suggest that weeping peach trees do not have defects in branch structure. Rather, transcriptomes from the adaxial (upper) and abaxial (lower) sides of standard and weeping branch shoot tips revealed flipped expression patterns for genes associated with early auxin response, tissue patterning, cell elongation, and tension wood development. This suggests that WEEP promotes polar auxin transport toward the lower side during shoot gravitropic response, leading to cell elongation and tension wood development. In addition, weeping peach trees exhibited steeper root systems and faster lateral root gravitropic response. This suggests that WEEP moderates root gravitropism and is essential to establishing the set-point angle of lateral roots from the gravity vector. Additionally, size-exclusion chromatography indicated that WEEP proteins self-oligomerize, like other proteins with sterile alpha motif (SAM) domains. Collectively, our results from weeping peach provide insight into polar auxin transport mechanisms associated with gravitropism and lateral shoot and root orientation."
Authors: Deborah Delmer, Richard A. Dixon, Kenneth Keegstra and Debra Mohnen.
The Plant Cell (2024)
One-sentence summary: This review provides a historical context for the processes by which plant cell wall structures are created, assemble, sense, and respond to signals and change during progression from birth to death.
Abstract: "Mythology is replete with good and evil shapeshifters, who, by definition, display great adaptability and assume many different forms—with several even turning themselves into trees. Cell walls certainly fit this definition as they can undergo subtle or dramatic changes in structure, assume many shapes, and perform many functions. In this review, we cover the evolution of knowledge of the structures, biosynthesis, and functions of the 5 major cell wall polymer types that range from deceptively simple to fiendishly complex. Along the way, we recognize some of the colorful historical figures who shaped cell wall research over the past 100 years. The shapeshifter analogy emerges more clearly as we examine the evolving proposals for how cell walls are constructed to allow growth while remaining strong, the complex signaling involved in maintaining cell wall integrity and defense against disease, and the ways cell walls adapt as they progress from birth, through growth to maturation, and in the end, often function long after cell death. We predict the next century of progress will include deciphering cell type–specific wall polymers; regulation at all levels of polymer production, crosslinks, and architecture; and how walls respond to developmental and environmental signals to drive plant success in diverse environments.
Authors: Haiqiang Zhang, Zichen Liu, Yunxiao Wang, Siyu Mu, Hongzhong Yue, Yanjie Luo, Zhengao Zhang, Yuhong Li and Peng Chen.
Theoretical and Applied Genetics (2024)
Key message A novel super compact mutant, scp-3, was identified using map-based cloning in cucumber. The CsDWF7 gene encoding a delta7 sterol C-5(6) desaturase was the candidate gene of scp-3.
Abstract: "Mining dwarf genes is important in understanding stem growth in crops. However, only a small number of dwarf genes have been cloned or characterized. Here, we characterized a cucumber (Cucumis sativus L.) dwarf mutant, super compact 3 (scp-3), which displays shortened internodes and dark green leaves with a wrinkled appearance. The photosynthetic rate of scp-3 is significantly lower than that of the wild type. The dwarf phenotype of scp-3 mutant can be partially rescued by the exogenous brassinolide (BL) application, and the endogenous brassinosteroids (BRs) levels in the scp-3 mutant were significantly lower compared to the wild type. Microscopic examination revealed that the reduced internode length in scp-3 resulted from a decrease in cell size. Genetic analysis showed that the dwarf phenotype of scp-3 was controlled by a single recessive gene. Combined with bulked segregant analysis and map-based cloning strategy, we delimited scp-3 locus into an 82.5 kb region harboring five putative genes, but only one non-synonymous mutation (A to T) was discovered between the mutant and its wild type in this region. This mutation occurred within the second exon of the CsGy4G017510 gene, leading to an amino acid alteration from Leu156 to His156. This gene encodes the CsDWF7 protein, an analog of the Arabidopsis DWF7 protein, which is known to be involved in the biosynthesis of BRs. The CsDWF7 protein was targeted to the cell membrane. In comparison to the wild type, scp-3 exhibited reduced CsDWF7 expression in different tissues. These findings imply that CsDWF7 is essential for both BR biosynthesis as well as growth and development of cucumber plants."
Authors: Ruo-Xi Zhang, Yudi Liu, Xian Zhang, Xiaomei Chen, Juanli Sun, Yun Zhao, Jinyun Zhang, Jia-Long Yao, Liao Liao, Hui Zhou and Yuepeng Han.
New Phytologist (2024)
Abstract: "Although maturity date (MD) is an essential factor affecting fresh fruit marketing and has a pleiotropic effect on fruit taste qualities, the underlying mechanisms remain largely unclear. In this study, we functionally characterized two adjacent NAM-ATAF1/2-CUC2 (NAC) transcription factors (TFs), PpNAC1 and PpNAC5, both of which were associated with fruit MD in peach. PpNAC1 and PpNAC5 were found capable of activating transcription of genes associated with cell elongation, cell wall degradation and ethylene biosynthesis, suggesting their regulatory roles in fruit enlargement and ripening. Furthermore, PpNAC1 and PpNAC5 had pleiotropic effects on fruit taste due to their ability to activate transcription of genes for sugar accumulation and organic acid degradation. Interestingly, both PpNAC1 and PpNAC5 orthologues were found in fruit-producing angiosperms and adjacently arranged in all 91 tested dicots but absent in fruitless gymnosperms, suggesting their important roles in fruit development. Our results provide insight into the regulatory roles of NAC TFs in MD and fruit taste."
Authors: Vishal Varshney and Manoj Majee.
Molecular Plant (2023)
Excerpts: "Recently, a study by Chen et al. (2023) delves into the role of the MKK3–MPK7–ERF4 module in controlling dormancy release and embryo expansion by regulating the expression of EXPAs. It provides insight into the awakening of dormant seeds by exploring three major aspects: (i) the involvement of the MKK3–MPK7 cascade, a crucial signaling pathway, in controlling seed dormancy; (ii) the elucidation of ERF4 as a substrate that negatively influences dormancy release, embryo expansion, and how degradation of ERF4 promotes dormancy release; and (iii) the role of EXPAs during imbibition and their responsiveness to dormancy-breaking factors (Chen et al., 2023)."
"In summary, the MKK3–MPK7–ERF4 module is proposed to be a critical “fate switch” governing the transition from seed dormancy to germination. The authors illustrate that EXPAs act as positive regulators for root cell expansion, and their expression is inhibited by ERF4. However, the cell-generated H2O2 activates the MKK3–MPK7 cascade that results in the phosphorylation of ERF4, leading to its subsequent degradation through the ubiquitin-proteasome pathway. The degradation of ERF4 releases this inhibition on EXPAs, leading to increased EXPA expression and, consequently, the promotion of root cell expansion in the transitional region near the radicle and lower hypocotyl, ultimately leading to the emergence of the radicle during seed imbibition (Figure 1)."
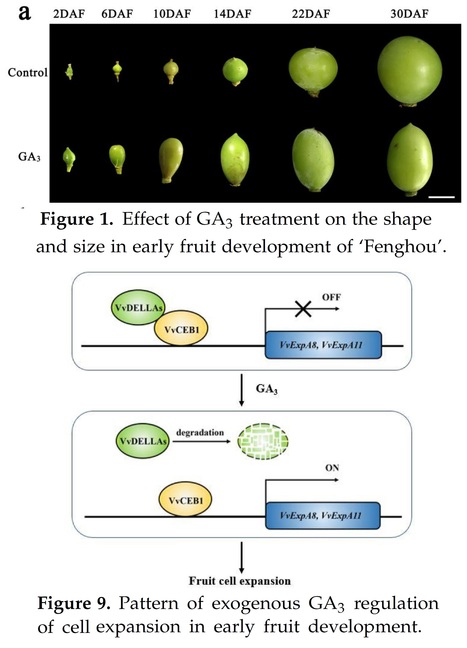
Authors: Zhenhua Liu, Yan Wang, Pingyin Guan, Jianfang Hu and Lei Sun.
International Journal of Molecular Sciences (2023)
Abstract: "Exogenous gibberellin treatment can promote early growth of grape fruit, but the underlying regulatory mechanisms are not well understood. Here, we show that VvDELLA2 directly regulates the activity of the VvCEB1 transcription factor, a key regulator in the control of cell expansion in grape fruit. Our results show that VvCEB1 binds directly to the promoters of cell expansion-related genes in grape fruit and acts as a transcriptional activator, while VvDELLA2 blocks VvCEB1 function by binding to its activating structural domain. The exogenous gibberellin treatment relieved this inhibition by promoting the degradation of VvDELLA2 protein, thus, allowing VvCEB1 to transcriptionally activate the expression of cell expansion-related genes. In conclusion, we conclude that exogenous GA3 treatment regulates early fruit expansion by affecting the VvDELLA-VvCEB1 interaction in grape fruit development."
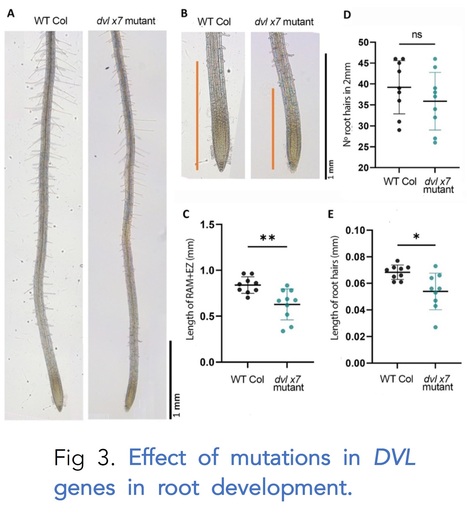
Authors: Ana Alarcia, Amparo Primo-Capella, Elena Perpiñán, Priscilla Rossetto and Cristina Ferrándiz.
bioRxiv (2023)
Abstract: "The DEVIL/ROTUNDIFOLIA-LIKE (DVL/RTFL) family of plant peptides is present in all land plants, but their biological role and mode of action remains largely unknown, in part due to the lack of reported phenotypes associated to DVL/RTFL loss of function. In this work we have generated high order mutants and characterized their phenotypes in reproductive development. Our results indicate that dvl mutants are affected in cell elongation processes, mainly related to pollen tube growth, and they appear to provide robustness to these processes. We also show that DVL peptides may act in different domains to those where the corresponding genes are transcribed, suggesting a putative role in the coordination of organ growth by participating in cell-to-cell communication."
Author: Elke Barbez.
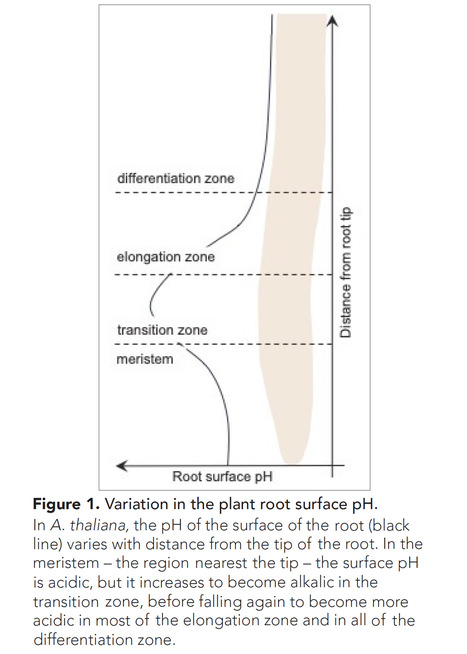
One-sentence summary: The growth of a plant root relies on careful control of root surface pH.
eLife (2023)
Excerpts: "Cell elongation is a crucial process in the growth of plants, both above and below ground. An old theory from the 1970s postulates that plant cells can stretch when the pH of the apoplast, the space outside of the cell membrane, is low (Rayle and Cleland, 1970). A plant hormone, called auxin, mediates this process by activating proton pumps in the cell membrane, leading to an increase in the number of protons into the apoplast and thus a lower pH. This in turn, activates specific enzymes that help to loosen the cell wall and thus cell expansion (Rayle and Cleland, 1970). In the following decades, scientists remained intrigued to unravel the molecular mechanism behind this ‘acid growth theory’. Now, in eLife, Matyas Fendrych and colleagues – including Nelson Serre and Daša Wernerová as joint first authors – report new insights into the role of pH in the growth of plant roots in the model organism Arabidopsis thaliana (Serre et al., 2023)."
"The regulation of membrane-based proton pumps is central in the acid growth theory. Serre et al. show, however, that the auxin-induced alkaline root zonation was not caused by altered proton pump activity, suggesting a distinct mode of action."
"In summary, Serre et al. illustrate that the root surface of A. thaliana features distinct pH zones, particularly highlighting the alkaline region in the transition zone. By identifying three molecular players in this process – AUX1, AFB1 and CNGC14 – they provide further molecular evidence to support the role of fast auxin responses in the pH-dependent regulation of root growth."
|


 Your new post is loading...
Your new post is loading...