Your new post is loading...
 Your new post is loading...
Authors: s Qi Wu, Jinyan Xu, Yingdi Zhao, Yuancong Wang, Ling Zhou, Lihua Ning, Sergey Shabala and Han Zhao.
Plant Physiology (2024)
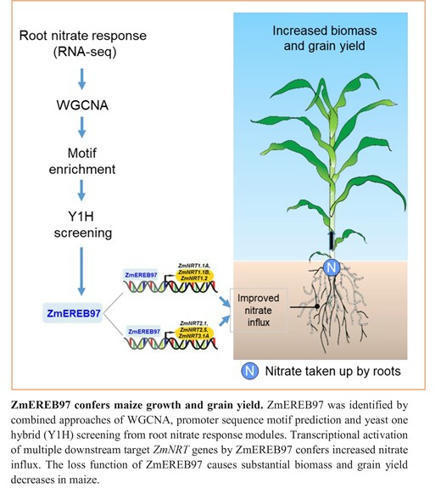
One-sentence summary: An ethylene response transcription factor plays an important role in the nitrogen-signaling network and in optimizing nitrate uptake in maize.
Abstract: "Maize (Zea mays L.) has very strong requirements for nitrogen. However, the molecular mechanisms underlying the regulations of nitrogen uptake and translocation in this species are not fully understood. Here, we report that an APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor ZmEREB97 functions as an important regulator in the N-signaling network in maize. Predominantly expressed and accumulated in main root and lateral root primordia, ZmEREB97 rapidly responded to nitrate treatment. By overlapping the analyses of differentially expressed genes and conducting a DAP-seq assay, we identified 1446 potential target genes of ZmEREB97. Among these, 764 genes were co-regulated in two lines of zmereb97 mutants. Loss of function of ZmEREB97 substantially weakened plant growth under both hydroponic and soil conditions. Physiological characterization of zmereb97 mutant plants demonstrated that reduced biomass and grain yield were both associated with reduced nitrate influx, decreased nitrate content and less N accumulation. We further demonstrated that ZmEREB97 directly targets and regulates the expression of six ZmNRT genes by binding to the GCC box-related sequences in gene promoters. Collectively, these data suggest that ZmEREB97 is a major positive regulator of the nitrate response and that it plays an important role in optimizing nitrate uptake, offering a target for improvement of nitrogen use efficiency in crops."
Authors: Vojtěch Schmidt, Roman Skokan, Thomas Depaepe, Katarina Kurtović, Samuel Haluška, Stanislav Vosolsobě, Roberta Vaculíková, Anthony Pil, Petre Ivanov Dobrev, Václav Motyka, Dominique Van Der Straeten and Jan Petrášek.
Nature Communications (2024)
Editor's view: Here, the authors show that the biosynthesis of many compounds in green algae preceded their recruitment in phytohormone signaling and metabolism in land plants.
Abstract: "The genomes of charophyte green algae, close relatives of land plants, typically do not show signs of developmental regulation by phytohormones. However, scattered reports of endogenous phytohormone production in these organisms exist. We performed a comprehensive analysis of multiple phytohormones in Viridiplantae, focusing mainly on charophytes. We show that auxin, salicylic acid, ethylene and tRNA-derived cytokinins including cis-zeatin are found ubiquitously in Viridiplantae. By contrast, land plants but not green algae contain the trans-zeatin type cytokinins as well as auxin and cytokinin conjugates. Charophytes occasionally produce jasmonates and abscisic acid, whereas the latter is detected consistently in land plants. Several phytohormones are excreted into the culture medium, including auxin by charophytes and cytokinins and salicylic acid by Viridiplantae in general. We note that the conservation of phytohormone biosynthesis and signaling pathways known from angiosperms does not match the capacity for phytohormone biosynthesis in Viridiplantae. Our phylogenetically guided analysis of established algal cultures provides an important insight into phytohormone biosynthesis and metabolism across Streptophyta. Genomic evidence dates the origins of most phytohormones to terrestrialization or later."
Authors: Tao Wang, Xuemin Ma, Ying Chen, Cuicui Wang, Zhenxiao Xia, Zixi Liu, Lihong Gao and Wenna Zhang.
Plant, Cell & Environment (2024)
Abstract: "Low temperature stress poses a significant challenge to the productivity of horticultural crops. The dynamic expression of cold-responsive genes plays a crucial role in plant cold tolerance. While NAC transcription factors have been extensively studied in plant growth and development, their involvement in regulating plant cold tolerance remains poorly understood. In this study, we focused on the identification and characterisation of SlNAC3 as the most rapid and robust responsive gene in tomato under low temperature conditions. Manipulating SlNAC3 through overexpression or silencing resulted in reduced or enhanced cold tolerance, respectively. Surprisingly, we discovered a negative correlation between the expression of CBF and cold tolerance in the SlNAC3 transgenic lines. These findings suggest that SlNAC3 regulates tomato cold tolerance likely through a CBF-independent pathway. Furthermore, we conducted additional investigations to identify the molecular mechanisms underlying SlNAC3-mediated cold tolerance in tomatoes. Our results revealed that SlNAC3 controls the transcription of ethylene biosynthetic genes, thereby bursting ethylene release in response to cold stress. Indeed, the silencing of these genes led to an augmentation in cold tolerance. This discovery provides valuable insights into the regulatory pathways involved in ethylene-mediated cold tolerance in tomatoes, offering potential strategies for developing innovative approaches to enhance cold stress resilience in this economically important crop species."
Authors: Sahana Basu, Monika, Surbhi Kumari and Gautam Kumar.
Plant Physiology and Biochemistry (2024)
Highlights: • IAA induces lateral root formation in rice under submergence • ROS and RNS cause lysigenous aerenchyma formation in root cortex of rice • Submergence decreases ABA level, causing GA-mediated escape response in rice shoot • Enhanced antioxidant levels dampen nitro-oxidative damage during de-submergence
Abstract: "Constant change in global climate has become the most important limiting factor to crop productivity. Asymmetrical precipitations are causing recurrent flood events around the world. Submergence is one of the most detrimental abiotic stresses for sustainable rice production in the rainfed ecosystems of Southeast Asia. Therefore, the development of submergence-tolerant rice is an essential requirement to encounter food security. Submergence tolerance in rice is governed by the major quantitative trait locus (QTL) designated as Submergence1 (Sub1) near the centromere of chromosome 9. The introduction of the Sub1 in high-yielding rice varieties producing near-isogenic lines (NILs) has shown extreme submergence tolerance. The present study aimed to understand the responses of rice genotype IR64 and its Sub1 NIL IR64 Sub1 following one week of complete submergence treatment. Submergence imposed severe nitro-oxidative stress in both the rice genotypes, consequently disrupting the cellular redox homeostasis. In this study, IR64 exhibited higher NADPH oxidase activity accompanied by increased reactive oxygen species, reactive nitrogen species, and malondialdehyde buildups and cell death under submergence. Higher accumulations of 1-Aminocyclopropane-1-carboxylic acid, gibberellic acid, and Indole-3-acetic acid were also observed in IR64 which accelerated the plant growth and root cortical aerenchyma development following submergence. In contrast, IR64 Sub1 had enhanced submergence tolerance associated with an improved antioxidant defense system with sustainable morpho-physiological activities and restricted root aerenchyma formation. The comprehensive analyses of the responses of rice genotypes with contrasting submergence tolerance may demonstrate the intricacies of rice under complete submergence and may potentially contribute to improving stress resilience by advancing our understanding of the mechanisms of submergence tolerance in rice."
Authors: Juncai Deng, Xiangqing Huang, Jianhua Chen, Bartel Vanholme, Jinya Guo, Yuanyuan He, Wenting Qin, Jing Zhang, Wenyu Yang and Jiang Liu.
Plant Physiology and Biochemistry (2024)
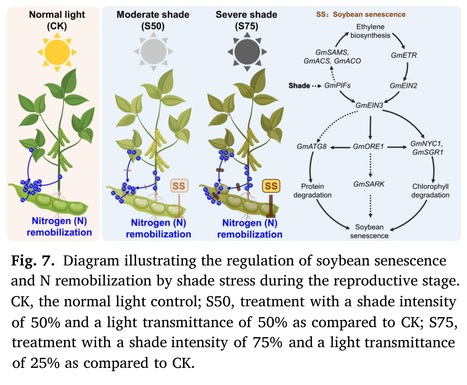
Highlights: • Shade stress leads to premature senescence in soybean plants. • Ethylene biosynthesis and signal transduction are induced to regulate soybean plant senescence. • Shade stress retains nitrogen in soybean vegetative organs and impedes the remobilization of nitrogen from these organs. • These negative impacts can be mitigated by reducing the intensity of shade stress through field layout optimization.
Abstract: "In gramineae-soybean intercropping systems, shade stress caused by taller plants impacts soybean growth specifically during the reproductive stage. However, the effects of shade stress on soybean senescence remain largely unexplored. In this research, we applied artificial shade treatments with intensities of 75% (S75) and 50% (S50) to soybean plants at the onset of flowering to simulate the shade stress experienced by soybeans in the traditional and optimized maize-soybean intercropping systems, respectively. Compared to the normal light control, both shade treatments led to a rapid decline in the dry matter content of soybean vegetative organs and accelerated their abscission. Moreover, shade treatments triggered the degradation of chlorophyll and soluble proteins in leaves and increased the expression of genes associated with leaf senescence. Metabolic profiling further revealed that ethylene biosynthesis and signal transduction were induced by shade treatment. In addition, the examination of nitrogen content demonstrated that shade treatments impeded the remobilization of nitrogen in vegetative tissues, consequently reducing the seed nitrogen harvest. It's worth noting that these negative effects were less pronounced under the S50 treatment compared to the S75 treatment. Taken together, this research demonstrates that shade stress during the reproductive stage accelerates soybean senescence and impedes nitrogen remobilization, while optimizing the field layout to improve soybean growth light conditions could mitigate these challenges in the maize-soybean intercropping system."
Authors: Tomoaki Sakamoto, Shuka Ikematsu, Hokuto Nakayama, Terezie Mandáková, Gholamreza Gohari, Takuya Sakamoto, Gaojie Li, Hongwei Hou, Sachihiro Matsunaga, Martin A. Lysak and Seisuke Kimura
Communications Biology (2024)
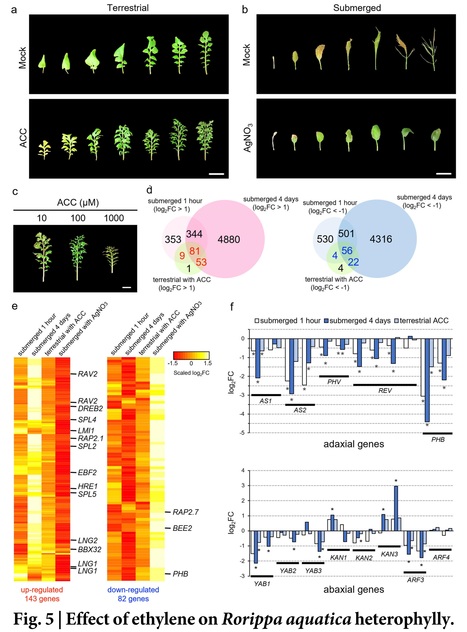
One-sentence summary: A chromosome-level genome assembly for the amphibious plant Rorippa aquatica uncovers its tetraploid origin and an involvement of ethylene in heterophylly upon submergence.
Abstract: "The ability to respond to varying environments is crucial for sessile organisms such as plants. The amphibious plant Rorippa aquatica exhibits a striking type of phenotypic plasticity known as heterophylly, a phenomenon in which leaf form is altered in response to environmental factors. However, the underlying molecular mechanisms of heterophylly are yet to be fully understood. To uncover the genetic basis and analyze the evolutionary processes driving heterophylly in R. aquatica, we assembled the chromosome-level genome of the species. Comparative chromosome painting and chromosomal genomics revealed that allopolyploidization and subsequent post-polyploid descending dysploidy occurred during the speciation of R. aquatica. Based on the obtained genomic data, the transcriptome analyses revealed that ethylene signaling plays a central role in regulating heterophylly under submerged conditions, with blue light signaling acting as an attenuator of ethylene signal. The assembled R. aquatica reference genome provides insights into the molecular mechanisms and evolution of heterophylly."
Authors: Chiara Pucciariello and Pierdomenico Perata.
Journal of Experimental Botany (2024)
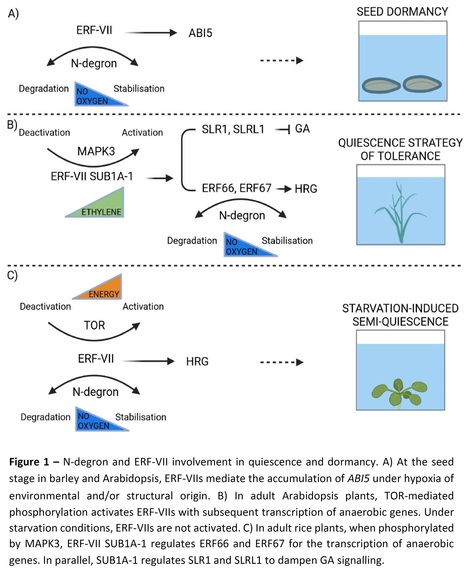
Abstract: "Plant quiescence and seed dormancy can be triggered by reduced oxygen availability. Under water, oxygen depletion caused by flooding can culminate in a quiescent state, which is a plant strategy for energy preservation and survival. In adult plants, a quiescent state can be activated by sugar starvation, culminating in metabolic depression. In seeds, secondary dormancy can be activated by reduced oxygen availability, which creates an unfavourable state for germination. The physical dormancy of some seeds and buds includes barriers to external conditions, which indirectly results in hypoxia. The molecular processes that support seed dormancy and plant survival through quiescence under hypoxia include the N-degron pathway, which enables the modulation of ethylene responsive factors of group VII and downstream targets. This oxygen- and nitric oxide-dependent mechanism interacts with phytohormone-related pathways to control growth."
Authors: Nguyen Hoai Nguyen, Phuong Thi Bich Ho and Linh Thi Truc Le.
Plant Reproduction (2024)
Key message: This review provides a thorough and comprehensive perspective on the topic of cucumber sexual expression. Specifically, insights into sex expression mediated by pathways other than ethylene are highlighted.
Abstract: "Cucumber (Cucumis sativus L.) is a common and important commercial crop that is cultivated and consumed worldwide. Additionally, this species is commonly used as a model for investigating plant sex expression. Cucumbers exhibit a variety of floral arrangements, comprising male, female, and hermaphroditic (bisexual) flowers. Generally, cucumber plants that produce female flowers are typically preferred due to their significant impact on the overall output. Various environmental conditions, such as temperature, light quality, and photoperiod, have been also shown to influence the sex expression in this species. Multiple lines of evidence indicate that ethylene and its biosynthesis genes are crucial in regulating cucumber sex expression. Gibberellins, another well-known phytohormone, can similarly influence cucumber sex expression via an ethylene-independent route. Further studies employing the next-generation sequencing technology also visualized a deeper slice of the molecular mechanism such as the role of the cell cycle program in the cucumber sex expression. This review aims to provide an overview of the sex expression of cucumber including its underlying molecular mechanism and regulatory aspects based on recent investigations."
Authors: Yuan-Chi Chien and Gyeong Mee Yoon.
BioEssays (2024)
Abstract: "Volatile compounds, such as nitric oxide and ethylene gas, play a vital role as signaling molecules in organisms. Ethylene is a plant hormone that regulates a wide range of plant growth, development, and responses to stress and is perceived by a family of ethylene receptors that localize in the endoplasmic reticulum. Constitutive Triple Response 1 (CTR1), a Raf-like protein kinase and a key negative regulator for ethylene responses, tethers to the ethylene receptors, but undergoes nuclear translocation upon activation of ethylene signaling. This ER-to-nucleus trafficking transforms CTR1 into a positive regulator for ethylene responses, significantly enhancing stress resilience to drought and salinity. The nuclear trafficking of CTR1 demonstrates that the spatiotemporal control of ethylene signaling is essential for stress adaptation. Understanding the mechanisms governing the spatiotemporal control of ethylene signaling elements is crucial for unraveling the system-level regulatory mechanisms that collectively fine-tune ethylene responses to optimize plant growth, development, and stress adaptation."
Authors: Minglei Zhao, Chun-Lin Shi and Jianguo Li.
Fruit Research (2024)
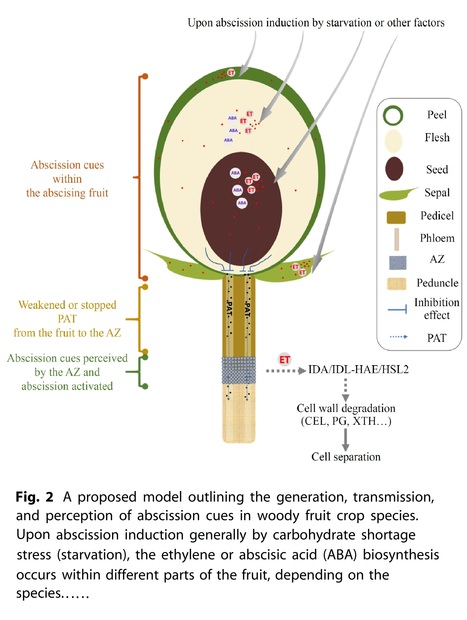
Abstract: "From an evolutionary perspective, fruit abscission is an intelligent regulatory mechanism by which fruit trees adapt to their environment and ensure offspring. However, from an agricultural production standpoint, unwanted fruit abscission can cause significant loss in fruit yield and economic value. Therefore, investigating the mechanisms of fruit abscission has always been an important focus in the field of plant research. Acquiring a thorough comprehension of the underlying mechanisms responsible for fruit abscission is highly valuable for enhancing fruit crop breeding and optimizing harvesting practices. In this review, we focus on fruit abscission, particularly discussing the nature of abscission cues within the abscising fruit, how these signals are generated and transmitted, and how the abscission zone cells perceive and respond to these signals in woody fruit crops."
Author: Aida Maric.
Plant Physiology (2024)
Excerpts: "Recent work by Kong et al. (2024), published in Plant Physiology reports ethylene-auxin hormonal crosstalk as a regulator of root angle in cereal crops. The authors used different root phenotyping methods to compare root response of wild-type and ethylene-insensitive rice and maize mutants. EIL1 and EIN2 are crucial links in the ethylene signaling pathway, conducting ethylene signal from the endoplasmic reticulum to the nucleus to control ethylene-responsive genes. Rice and maize mutants of EIL1 and EIN2 (oseil1, osein2 and zmein2-1) were not able to sense ethylene signals and had impaired ethylene response. Kong et al. (2024) showed that oseil1 and osein2 mutants have a shallower root system with larger root angles (Figure 1A). Additionally, ethylene mutants had an impaired gravitropic response compared to wild-type plants."
"By comparing hormone profiles of ethylene-insensitive mutants with wild-type plants they showed that ethylene-insensitive mutants exhibit lower levels of auxin. Exogenous application of auxin could rescue the root angle phenotype of the ethylene-insensitive mutants (Figure 1B), pinpointing auxin as the intermediate signal in ethylene-mediated root growth angles. Furthermore the authors showed that MHZ10, a member of the auxin biosynthesis pathway, plays a key role in the ethylene-mediated regulation of root angle (Figure 1B)."
Authors: Mingjuan Zhai, Yating Chen, Xiaowu Pan, Ying Chen, Jiahao Zhou, Xiaodan Jiang, Zhijin Zhang, Guiqing Xiao and Haiwen Zhang.
Plant, Cell & Environment (2024)
One-sentence summary: The OsEIN2-OsEIL1/2 ethylene pathway has negative effect on rice tolerance to low temperature stress through transcriptionally repressing OsICE1-mediated chilling response.
Abstract: "Low temperature severely affects rice development and yield. Ethylene signal is essential for plant development and stress response. Here, we reported that the OsEIN2-OsEIL1/2 pathway reduced OsICE1-dependent chilling tolerance in rice. The overexpressing plants of OsEIN2, OsEIL1 and OsEIL2 exhibited severe stress symptoms with excessive reactive oxygen species (ROS) accumulation under chilling, while the mutants (osein2 and oseil1) and OsEIL2-RNA interference plants (OsEIL2-Ri) showed the enhanced chilling tolerance. We validated that OsEIL1 and OsEIL2 could form a heterodimer and synergistically repressed OsICE1 expression by binding to its promoter. The expression of OsICE1 target genes, ROS scavenging- and photosynthesis-related genes were downregulated by OsEIN2 and OsEIL1/2, which were activated by OsICE1, suggesting that OsEIN2-OsEIL1/2 pathway might mediate ROS accumulation and photosynthetic capacity under chilling by attenuating OsICE1 function. Moreover, the association analysis of the seedling chilling tolerance with the haplotype showed that the lower expression of OsEIL1 and OsEIL2 caused by natural variation might confer chilling tolerance on rice seedlings. Finally, we generated OsEIL2-edited rice with an enhanced chilling tolerance. Taken together, our findings reveal a possible mechanism integrating OsEIN2-OsEIL1/2 pathway with OsICE1-dependent cascade in regulating chilling tolerance, providing a practical strategy for breeding chilling-tolerant rice."
Authors: Jing Chen, Senlin Jiang, Guobin Yang, Lujun Li, Jing Li and Fengjuan Yang.
Plant Physiology and Biochemistry (2024)
Highlights: • SmMYB113 can increase flower drop rate in eggplant. • SmMYB113 regulates ethylene-dependent flower abscission by directly activating the ethylene biosynthetic genes. • SmERF38 enhanced the activation of SmMYB113 on the promoters of SmACS1 by interaction.
Abstract: "Flower abscission is an important developmental process that can significantly reduce the yield of horticultural plants. We previously reported that SmMYB113 is a key transcription factor promoting anthocyanin biosynthesis and improve fruit quality. However, the overexpression of SmMYB113 in eggplant increased flower drop rate and reduced fruit yield. Here, we elucidate the regulatory mechanisms of SmMYB113 on flower abscission in eggplant. RNA-seq analysis indicated that the regulation of flower abscission by SmMYB113 was associated with altered expression of genes related to ethylene biosynthesis and signal transduction, including ethylene biosynthetic genes SmACS1, SmACS8 and SmACO4. Then, the ethylene content in flowers and the function of ethephon (ETH, which promotes fruit ripening) and 1-Methylcyclopropene (1-MCP, which acts as an ethylene perception inhibitor) were analyzed, which revealed that SmMYB113 directly regulates ethylene-dependent flower abscission. Yeast one-hybrid and dual-luciferase assays revealed that SmMYB113 could directly bind to the promoters of SmACS1, SmACS8, and SmACO4 to activate their expression. Through construction of a yeast two-hybrid (Y2H) screening library, the protein SmERF38 was found to interact with SmMYB113, and verified by Y2H, bimolecular fluorescence complementation (BiFC), and luciferase complementation assay. Furthermore, dual-luciferase assays showed that SmERF38 enhanced the role of SmMYB113 on the promoters of SmACS1. Our results provided new insight into the molecular mechanism of flower abscission in eggplant."
|
Authors: Huixin Chen, Dongdong Li and Kunsong Chen.
Fruit Research (2024)
Abstract: "Strawberry, considered to be a model for non-climacteric fruits, has traditionally been viewed as having a less pronounced reliance on ethylene in fruit development. However, the inherent tissue heterogeneity within strawberry fruit, coupled with variable ethylene production levels, suggests a potential role for ethylene in the maturation of the true fruit, the achenes, dispersed on the fleshy receptacle. This intricate process is likely to exert ripple effects on the subsequent growth and ripening of the receptacle. To comprehensively unravel the functions of ethylene in strawberry fruit, there is a need for ethylene detection sensors, ethylene response reporter transgenic plants, and genetically engineered mutants achieved through genome editing, encompassing both biosynthesis and signaling mutants."
Authors: Dongmei Yan and Huilan Yi.
Scientia Horticulturae (2024)
Highlights • Identifying key genes and pathways for postharvest preservation of grapes by RNA-seq. • SO2 prolongs the postharvest shelf life of table grapes by reducing cell wall degradation and enhancing defenses of the skins. • SO2-triggered secondary metabolism and defense responses in grape fruits contributed to postharvest preservation. • SO2-induced inhibition of ethylene signaling pathway helped to delay postharvest softening and aging of grape fruits.
Abstract: "Sulfur dioxide (SO2) is the most frequently used preservative for table grapes, yet the preservation mechanism remains unclear. To ascertain the specific genes and pathways involved in SO2 preservation, RNA-seq technology was employed to characterize the transcriptome profile of grapes during postharvest storage. A total of 22,288 genes were identified, of which 377 genes were differentially expressed (≥ 2-fold change) between SO2 and control groups, mainly enriched in secondary metabolism, plant-pathogen interactions, plant hormone signaling, etc. Numerous genes encoding pathogenesis-related (PR) proteins exhibited higher expression levels in SO2 group, while the activities of disease-resistant enzymes, such as β-1,3-glucanase (PR2) and chitinase (PR3) significantly increased by 28.9 % and 29.3 % (P < 0.05), indicating SO2-induced plant resistance. Exposure to SO2 also enhanced the expression of genes encoding essential enzymes of secondary metabolism, and significantly increased both the activities of critical enzymes for the biosynthesis of secondary metabolites, such as phenylalanine ammonia-lyase (PAL) and 4-coumarate-CoA ligase (4CL), and the contents of secondary metabolites such as total phenol, flavonoid, anthocyanin, and lignin, demonstrating an enhanced chemical and physical barriers in SO2-fumigated grapes. Meanwhile, the genes associated with ethylene signaling and cell wall degradation were down-regulated by SO2 preservative, and some ethylene-responsive elements were identified in the promoter regions of cell wall hydrolase genes, suggesting that SO2-inhibited ethylene signaling might contribute to maintaining cell wall integrity. Altogether, gene expression patterns and cellular physiological processes were altered in grapes exposed to SO2, which helped maintain fruit quality and prolong postharvest life by regulating the defense responses and fruit ageing."
Authors: Wenli Hu, Rong Wang, Xiaohua Hao, Shaozhuang Li, Xinjie Zhao, Zijing Xie, Sha Wu, Liqun Huang, Ying Tan, Lianfu Tian and Dongping Li.
The Plant Journal (2024)
Significant Statement: The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight.
Abstract: "A fundamental question in developmental biology is how to regulate grain size to improve crop yields. Despite this, little is still known about the genetics and molecular mechanisms regulating grain size in crops. Here, we provide evidence that a putative protein kinase-like (OsLCD3) interacts with the S-adenosyl-L-methionine synthetase 1 (OsSAMS1) and determines the size and weight of grains. OsLCD3 mutation (lcd3) significantly increased grain size and weight by promoting cell expansion in spikelet hull, whereas its overexpression caused negative effects, suggesting that grain size was negatively regulated by OsLCD3. Importantly, lcd3 and OsSAMS1 overexpression (SAM1OE) led to large and heavy grains, with increased ethylene and decreased polyamines production. Based on genetic analyses, it appears that OsLCD3 and OsSAMS1 control rice grain size in part by ethylene/polyamine homeostasis. The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight."
Authors: Yanyan Tang, Zhong Huang, Shaohui Xu, Wenjie Zhou, Jianjun Ren, Fuxin Yu, Jingshan Wang, Wujun Ma and Lixian Qiao.
The Crop Journal (2024)
Abstract: "Ethylene plays essential roles in plant growth, development and stress responses. The ethylene signaling pathway and molecular mechanism have been studied extensively in Arabidopsis and rice but limited in peanuts. Here, we established a sand-culture method to screen pingyangmycin mutagenized peanut lines based on their specific response to ethylene (“triple response“). An ethylene-insensitive mutant, inhibition of peanut hypocotyl elongation 1 (iph1), was identified that showed reduced sensitivity to ethylene in both hypocotyl elongation and root growth. Through bulked segregant analysis sequencing, a major gene related to iph1, named AhIPH1, was preliminarily mapped at the chromosome Arahy.01, and further narrowed to a 450-kb genomic region through substitution mapping strategy. A total of 7014 genes were differentially expressed among the ACC treatment through RNA-seq analysis, of which only the Arahy.5BLU0Q gene in the candidate mapping interval was differentially expressed between WT and mutant iph1. Integrating sequence variations, functional annotation and transcriptome analysis revealed that a predicated gene, Arahy.5BLU0Q, encoding SNF1 protein kinase, may be the candidate gene for AhIPH1. This gene contained two single-nucleotide polymorphisms at promoter region and was more highly expressed in iph1 than WT. Our findings reveal a novel ethylene-responsive gene, which provides a theoretical foundation and new genetic resources for the mechanism of ethylene signaling in peanuts"
Authors: Xiaolin Cai, Wenyao Zhang, Jinnan Luo, Wei Li, Rui-hong Chen, Xiang-nan Xu, Ying-qiang Wen, Jia-yue Feng.
bioRxiv (2024)
Abstract: "Xanthomonas fragariae (Xaf) is the cause for strawberry crown dry cavity rot and strawberry leaf angular spots. Despite having a long evolutionary history with strawberries, the plant-pathogen connection is poorly understood. Pathogenicity for the majority of plant pathogens is mostly dependent on the type-III secretion system, which introduces virulence type III effectors (T3Es) into eukaryotic hosts cells. For most of these T3Es, the subcellular targets are yet unclear. Here, we used the yeast-two-hybrid (Y2H) technique to construct an interaction network of strawberry-Xaf T3Es. Multiple T3Es were discovered to converge onto the strawberry 1-aminocyclopropane-1-carboxylic acid oxidases (ACOs), which are the last rate-limited step in the production of ethylene. We then concentrated on the connection between XopL and FveACO9. Strawberry plants that overexpressed XopL accumulated higher levels of ethylene and exhibited more severe Xaf infection. XopL boosted ethylene production by stabilizing the accumulation of FveACO9 protein and enhancing ACO enzyme activity. Additionally, strawberries treated with ACC or overexpressing FveACO9 were particularly vulnerable to Xaf infection. On the other hand, pre-treatment with α-aminoinoisobutyric acid (AIB), an ACO inhibitor, effectively reduced Xaf infection. Our research indicates that Xaf utilizes a distinct approach to regulate the ethylene production of host plants in order to promote infection."
Authors: Lijuan Zhu, Haitao Yu, Xiaoyu Xu and Zhifang Yu.
Postharvest Biology and Technology (2024)
Highlights: • 10 μM MeJA inhibited ethylene biosynthesis and maintained better fruit quality in peach. • JA-biosynthetic pathway enhanced by MeJA was repressed during storage. • MeJA activated the negative feedback of JA-signaling pathway in peach.
Abstract: "Peach undergoes a rapid ripening and senescence, resulting in quality deterioration after harvest. Ethylene is essential for ripening and senescence in climacteric fruit. Although methyl jasmonate (MeJA) has been found to suppress ethylene biosynthesis, the underlying mechanism is unknown. We investigated the regulation of MeJA treatment (10 μM, 24 h) on the quality, ethylene biosynthesis and signaling, and jasmonic acid (JA) metabolism and signaling in ‘Xiahui 8’ peach stored at 20 °C. MeJA treatment maintained better fruit quality and reduced ethylene production (the reduction of peak value reached 24 %) during storage. The activities of 1-aminocyclopropane-1-carboxylate synthase (ACS) and 1-aminocyclopropane-1-carboxylate oxidase (ACO) decreased by 15–28 % and 15–19 % in MeJA treated fruit during storage. Additionally, the expressions of genes involved in ethylene signaling were downregulated in MeJA treated fruit. MeJA treatment enhanced JA biosynthesis through increasing the activities of allene oxide synthase (AOS), allene oxide cyclase (AOC) and JA-amino synthetase (JAR) during early storage. However, the JA-inducible catabolic pathways and self-repression of myelocytomatosis proteins2 (MYC2) were activated in MeJA treated fruit thereafter. This resulted in the decrease of PpMYC2 transcription and JA-Ile accumulation, which reduced by at most 56 % and 73 % when compared with CK, respectively, on day 0 and day 5. These results suggested that negative feedback regulation terminated and repressed the JA signaling, therefore diminishing its enhancement effect on ethylene biosynthesis in peach during storage."
Authors: Zhichao Sun, Xinmiao Guo, R.M. Saravana Kumar, Chunying Huang, Yan Xie, Meng Li and Jisheng Li.
Plant Science (2024)
Highlights: • Late stage of mulberry fruit ripening witnessed ethylene accumulation and physiological changes. • Late stage fruit ripening-specific differentially expressed genes and associated metabolic pathways highlighted. • Transcriptome and metabolome data interconnects ethylene signaling and mulberry fruit ripening. • MaERF3 are important for ripening process.
Abstract: "Mulberry (Morus alba L.) is a climacteric and highly perishable fruit. Ethylene has been considered to be an important trigger of fruit ripening process. However, the role of ethylene in the mulberry fruit ripening process remains unclear. In this study, we performed a comprehensive analysis of metabolomic and transcriptomic data of mulberry fruit and the physiological changes accompanying the fruit ripening process. Our study revealed that changes in the accumulation of specific metabolites at different stages of fruit development and ripening were closely correlated to transcriptional changes as well as underlying physiological changes and the development of taste biomolecules. The ripening of mulberry fruits was highly associated with the production of endogenous ethylene, and further application of exogenous ethylene assisted the ripening process. Transcriptomic analysis revealed that differential expression of diverse ripening-related genes was involved in sugar metabolism, anthocyanin biosynthesis, and cell wall modification pathways. Network analysis of transcriptomics and metabolomics data revealed that many transcription factors and ripening-related genes were involved, among which ethylene-responsive transcription factor 3 (MaERF3) plays a crucial role in the ripening process. The role of MaERF3 in ripening was experimentally proven in a transient overexpression assay in apples. Our study indicates that ethylene plays a vital role in modulating mulberry fruit ripening. The results provide a basis for guiding the genetic manipulation of mulberry fruits towards sustainable agricultural practices and improve post-harvest management, potentially enhancing the quality and shelf life of mulberry fruits for sustainable agriculture and forestry."
Authors: Xiaofang Li, Xin Zheng, Nikita Yadav, Shouvik Saha, El-Sayed Salama, Xiangkai Li, Likun Wang and Byong-Hun Jeon.
Plant Communications (2024)
Abstract: The Green Revolution of the mid-20th century transformed agriculture worldwide and has resulted in environmental challenges. A new approach, the Second Green Revolution, seeks to enhance agricultural productivity while minimizing negative environmental impacts. Plant microbiomes play critical roles in plant growth and stress responses, and understanding plant–microbiome interactions is essential for developing sustainable agricultural practices that meet food security and safety challenges, which are among the United Nations Sustainable Development Goals. This review provides a comprehensive exploration of key deterministic processes crucial for developing microbiome management strategies, including the host effect, the facilitator effect, and microbe–microbe interactions. A hierarchical framework for plant microbiome modulation is proposed to bridge the gap between basic research and agricultural applications. This framework emphasizes three levels of modulation: single-strain, synthetic community, and in situ microbiome modulation. Overall, rational management of plant microbiomes has wide-ranging applications in agriculture and can potentially be a core technology for the Second Green Revolution."
Authors: Xingshuai Ma, Zidi He, Ye Yuan, Zhijian Liang, Hang Zhang, Vilde Olsson Lalun, Zhuoyi Liu, Yanqing Zhang, Zhiqiang Huang, Yulian Huang, Jianguo Li and Minglei Zhao.
Journal of Integrative Plant Biology (2024)
Abstract: "At the physiological level, the interplay between auxin and ethylene has long been recognized as crucial for the regulation of organ abscission in plants. However, the underlying molecular mechanisms remain unknown. Here, we identified transcription factors involved in indoleacetic acid (IAA) and ethylene (ET) signaling that directly regulate the expression of INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and its receptor HAESA (HAE), which are key components initiating abscission. Specifically, litchi IDA-like 1 (LcIDL1) interacts with the receptor HAESA-like 2 (LcHSL2). Through in vitro and in vivo experiments, we determined that the auxin response factor LcARF5 directly binds and activates both LcIDL1 and LcHSL2. Furthermore, we found that the ETHYLENE INSENSITIVE 3-like transcription factor LcEIL3 directly binds and activates LcIDL1. The expression of IDA and HSL2 homologs was enhanced in LcARF5 and LcEIL3 transgenic Arabidopsis plants, but reduced in ein3 eil1 mutants. Consistently, the expressions of LcIDL1 and LcHSL2 were significantly decreased in LcARF5- and LcEIL3-silenced fruitlet abscission zones (FAZ), which correlated with a lower rate of fruitlet abscission. Depletion of auxin led to an increase in 1-aminocyclopropane-1-carboxylic acid (the precursor of ethylene) levels in the litchi FAZ, followed by abscission activation. Throughout this process, LcARF5 and LcEIL3 were induced in the FAZ. Collectively, our findings suggest that the molecular interactions between litchi AUXIN RESPONSE FACTOR 5 (LcARF5)–LcIDL1/LcHSL2 and LcEIL3–LcIDL1 signaling modules play a role in regulating fruitlet abscission in litchi and provide a long-sought mechanistic explanation for how the interplay between auxin and ethylene is translated into the molecular events that initiate abscission."
Authors: Kangning Zhang, Hongli Xie, Jiangqi Wen, Jing Zhang, Zeng-Yu Wang, Bin Xu and Maofeng Chai.
Grass Research (2024)
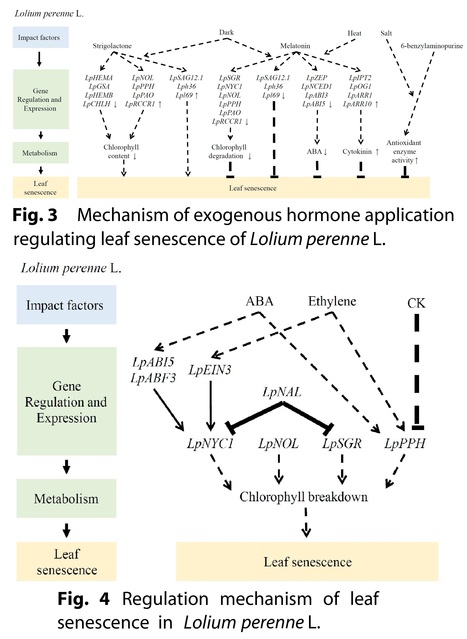
Abstract: "Leaf senescence is a complex biological process regulated by development, phytohormones, and various environmental factors. For forage and turf grasses, controlling leaf senescence can greatly improve forage quality, the amenity of lawn and turf, and the grasses’ stress tolerances. Leaf senescence involves a multitude of gene regulation and metabolic changes, including the alteration of chlorophyll metabolism. Here, we summarized the recent progress of studies on leaf senescence in major forage and turf grass species, such as Medicago truncatula, M. sativa, Lolium perenne, Panicum virgatum, and Agrostis stolonifera, to provide an insight into the development of effective methods for delaying leaf senescence in grass species."
Authors: James Giovannoni, Yao Chen, Xin Wang, Vincent Colantonio, Tara Fish, Jie Ye, Theodore Thannhauser, Zhibiao Ye, Mingchun Liu, Yongsheng Liu and Zhangjun Fei.
Research Square (2024)
Abstract: "Ripening is crucial for the development of fleshy fruits that release their seeds following consumption by frugivores and are important contributors to human health and nutritional security. Many genetic ripening regulators have been identified, especially in the model system tomato, yet more remain to be discovered and integrated into comprehensive regulatory models. Most tomato ripening genes have been studied in pericarp tissue, though recent evidence indicates that locule tissue is a site of early ripening-gene activities. Here we identified and functionally characterized an Ethylene Response Factor gene, SlERF.D6, by investigating tomato transcriptome data throughout plant development, emphasizing genes elevated in the locule during fruit development and ripening. SlERF.D6loss-of-function mutants resulting from CRISPR/Cas9 gene editing delayed ripening initiation and carotenoid accumulation in both pericarp and locule tissues. Transcriptome analysis of lines altered in SlERF.D6 expression revealed multiple classes of altered genes including ripening regulators, in addition to carotenoid, cell wall and ethylene pathway genes, suggesting comprehensive ripening control. Distinct regulatory patterns in pericarp versus locule tissues were observed indicating tissue-specific activity of this transcription factor. Analysis of SlERF.D6 interaction with target promoters revealed an AP2/ERF transcription factor (SlDEAR2) as a target of SlERF.D6. Furthermore, we show that a third transcription factor gene, SlTCP12, is a target of SlDEAR2, presenting a tri-component module of ripening control."
Author: Gwendolyn K. Kirschner.
The Plant Cell (2024)
Excerpts: "Yuxiang Li, Juan Wang and colleagues (Li et al. 2024) now connect the two roles of ethylene: as a signal responding to soil compaction and to trigger crown root development. In this work, they mimicked different levels of compaction by increasing agar concentrations and compared the growth of wildtype plants and ethylene signaling mutants. They found that compaction triggered crown root initiation via an ethylene-dependent pathway (Figure, A), and manipulating ethylene-related regulators influenced both root development and grain yield, implying that these regulators balance both root and grain development."
"By analyzing crown root numbers and transcript levels in oswox11 and ethylene signaling mutants, or overexpression combinations under normal or compacted soil conditions, the authors confirmed that the ethylene-OsEIL1-OsWOX11 module facilitates crown root development in compacted soil (Figure, B)."
|


 Your new post is loading...
Your new post is loading...