Your new post is loading...
 Your new post is loading...
Authors: Amanpreet Kaur, Norman B. Best, Thomas Hartwig, Josh Budka, Rajdeep Khangura, Steven McKenzie, Alejandro Aragón-Raygoza, Josh Strable, Burkhard Schulz and Brian P. Dilkes.
Plant Physiology (2023)
One-sentence summary: Molecular identity of a maize semidwarf mutant reveals a role for the maize GRAS domain transcription factor ortholog of DWARF AND LOW TILLERING in brassinosteroid signaling.
Abstract: "Brassinosteroids (BR) and gibberellins (GA) regulate plant height and leaf angle in maize (Zea mays). Mutants with defects in BR or GA biosynthesis or signaling identify components of these pathways and enhance our knowledge about plant growth and development. In this study, we characterized three recessive mutant alleles of GRAS transcription factor 42 (gras42) in maize, a GRAS transcription factor gene orthologous to the DWARF AND LOW TILLERING (DLT) gene of rice (Oryza sativa). These maize mutants exhibited semi-dwarf stature, shorter and wider leaves, and more upright leaf angle. Transcriptome analysis revealed a role for GRAS42 as a determinant of BR signaling. Analysis of the expression consequences from loss of GRAS42 in the gras42-mu1021149 mutant indicated a weak loss of BR signaling in the mutant, consistent with its previously demonstrated role in BR signaling in rice. Loss of BR signaling was also evident by the enhancement of weak BR biosynthetic mutant alleles in double mutants of nana plant1-1 and gras42-mu1021149. The gras42-mu1021149 mutant had little effect on GA-regulated gene expression, suggesting that GRAS42 is not a regulator of core GA signaling genes in maize. Single cell expression data identified gras42 expressed among cells in the G2/M phase of the cell cycle consistent with its previously demonstrated role in cell cycle gene expression in Arabidopsis (Arabidopsis thaliana). Cis-acting natural variation controlling GRAS42 transcript accumulation was identified by expression genome-wide association study (eGWAS) in maize. Our results demonstrate a conserved role for GRAS42/SCARECROW-LIKE 28 (SCL28)/DLT in BR signaling, clarify the role of this gene in GA signaling, and suggest mechanisms of tillering and leaf angle control by BR."
Authors: Tomoaki Sakamoto, Shuka Ikematsu, Hokuto Nakayama, Terezie Mandáková, Gholamreza Gohari, Takuya Sakamoto, Gaojie Li, Hongwei Hou, Sachihiro Matsunaga, Martin A. Lysak and Seisuke Kimura
Communications Biology (2024)
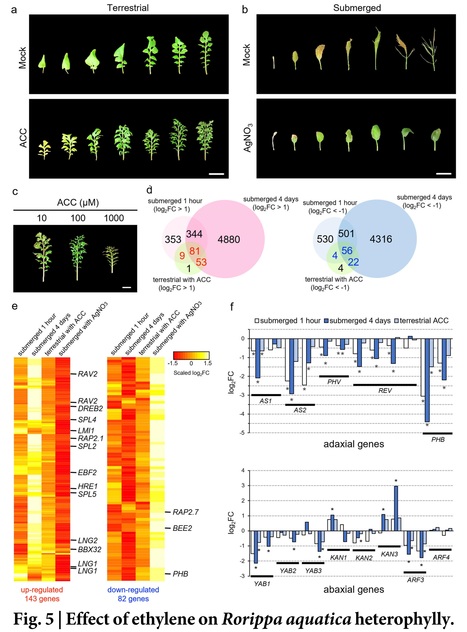
One-sentence summary: A chromosome-level genome assembly for the amphibious plant Rorippa aquatica uncovers its tetraploid origin and an involvement of ethylene in heterophylly upon submergence.
Abstract: "The ability to respond to varying environments is crucial for sessile organisms such as plants. The amphibious plant Rorippa aquatica exhibits a striking type of phenotypic plasticity known as heterophylly, a phenomenon in which leaf form is altered in response to environmental factors. However, the underlying molecular mechanisms of heterophylly are yet to be fully understood. To uncover the genetic basis and analyze the evolutionary processes driving heterophylly in R. aquatica, we assembled the chromosome-level genome of the species. Comparative chromosome painting and chromosomal genomics revealed that allopolyploidization and subsequent post-polyploid descending dysploidy occurred during the speciation of R. aquatica. Based on the obtained genomic data, the transcriptome analyses revealed that ethylene signaling plays a central role in regulating heterophylly under submerged conditions, with blue light signaling acting as an attenuator of ethylene signal. The assembled R. aquatica reference genome provides insights into the molecular mechanisms and evolution of heterophylly."
Authors: Daniela Vlad, Maricris Zaidem, Chiara Perico, Olga Sedelnikova, Samik Bhattacharya and Jane A. Langdale.
Current Biology (2024)
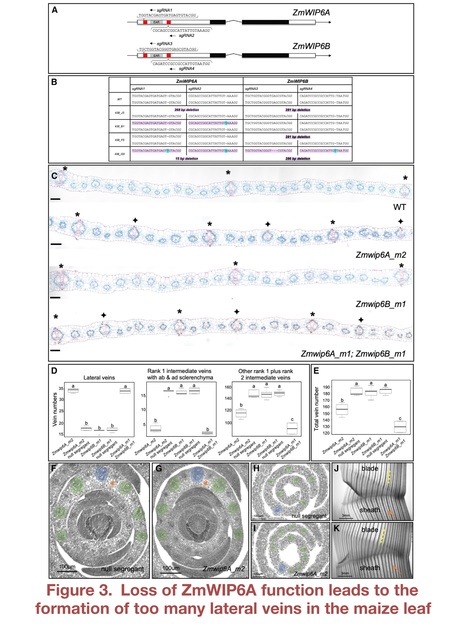
Editor's view: Vlad et al. discover a conserved mechanism that determines whether procambial stem cells in grass leaf primordia will develop into large lateral or smaller intermediate veins. The mechanism is revealed through the analysis of mutants defective in the activity of a zinc finger transcription factor of the WIP6 family that is named TOO MANY LATERALS.
Highlights • Function of WIP6 ortholog TML is conserved in maize and rice • TML is expressed in procambial cells during early leaf vein development • tml mutants develop large lateral veins in places where smaller veins should form • TML function suppresses lateral vein development in a subset of procambial initials
Abstract: "Grass leaves are invariantly strap shaped with an elongated distal blade and a proximal sheath that wraps around the stem. Underpinning this shape is a scaffold of leaf veins, most of which extend in parallel along the proximo-distal leaf axis. Differences between species are apparent both in the vein types that develop and in the distance between veins across the medio-lateral leaf axis. A prominent engineering goal is to increase vein density in leaves of C3 photosynthesizing species to facilitate the introduction of the more efficient C4 pathway. Here, we discover that the WIP6 transcription factor TOO MANY LATERALS (TML) specifies vein rank in both maize (C4) and rice (C3). Loss-of-function tml mutations cause large lateral veins to develop in positions normally occupied by smaller intermediate veins, and TML transcript localization in wild-type leaves is consistent with a role in suppressing lateral vein development in procambial cells that form intermediate veins. Attempts to manipulate TML function in rice were unsuccessful because transgene expression was silenced, suggesting that precise TML expression is essential for shoot viability. This finding may reflect the need to prevent the inappropriate activation of downstream targets or, given that transcriptome analysis revealed altered cytokinin and auxin signaling profiles in maize tml mutants, the need to prevent local or general hormonal imbalances. Importantly, rice tml mutants display an increased occupancy of veins in the leaf, providing a step toward an anatomical chassis for C4 engineering. Collectively, a conserved mechanism of vein rank specification in grass leaves has been revealed.
Authors: Jiacai Chen, Liu Liu, Guangxin Chen, Shaoyun Wang, Ye Liu, Zeqin Zhang, Hongfei Li, Liming Wang, Zhaoyang Zhou, Jianyu Zhao and Xiaolan Zhang.
Journal of Integrative Plant Biology (2024)
Abstract: "Leaves are the main photosynthesis organ that directly determines crop yield and biomass. Dissecting the regulatory mechanism of leaf development is crucial for food security and ecosystem turn-over. Here, we identified the novel function of R2R3-MYB transcription factors CsRAXs in regulating cucumber leaf size and fruiting ability. Csrax5 single mutant exhibited enlarged leaf size and stem diameter, and Csrax1/2/5 triple mutant displayed further enlargement phenotype. Overexpression of CsRAX1 or CsRAX5 gave rise to smaller leaf and thinner stem. The fruiting ability of Csrax1/2/5 plants was significantly enhanced, while that of CsRAX5 overexpression lines was greatly weakened. Similarly, cell number and free auxin level were elevated in mutant plants while decreased in overexpression lines. Biochemical data indicated that CsRAX1/5 directly promoted the expression of auxin glucosyltransferase gene CsUGT74E2. Therefore, our data suggested that CsRAXs function as repressors for leaf size development by promoting auxin glycosylation to decrease free auxin level and cell division in cucumber. Our findings provide new gene targets for cucumber breeding with increased leaf size and crop yield."
Author: Pablo Ignacio Calzadilla.
Plant Physiology (2024)
Excerpts: "In this Issue of Plant Physiology, Chang et al. (2024) studied the connection between NAC TFs and JA in regulating leaf senescence in tobacco plants. Exploring previous transcriptome data, the authors identified NtNAC56 as a candidate regulatory TF for leaf senescence. To study NtNAC56’s role, transgenic lines over-expressing NtNAC56 (NtNAC56-OE) were generated and characterized. The NtNAC56-OE lines senesced earlier than the wild type (WT) plants. NtNAC56-OE plants had significantly lower chlorophyll content and photosynthetic capacity and higher expression of SAGs when compared to WT."
"In summary, Chang et al. (2024) used a broad variety of techniques to explore the regulatory mechanisms inducing leaf senescence in tobacco. They identified NtNAC56 as a novel TF contributing to JA-induced leaf senescence. Data showed a positive activation loop between NtNAC56 and JA, and that NtNAC56 directly regulates JA biosynthesis through its binding to the specific TTTCTT cis-regulatory motif (Figure 1). Overall, this study contributes to our knowledge of the complex signaling network regulating leaf senescence."
Authors: Vijay Pratap Singh, Saumya Jaiswal, Yuanyuan Wang, Shouli Feng, Durgesh Kumar Tripathi, Samiksha Singh, Ravi Gupta, Dawei Xue, Shengchun Xu and Zhong-Hua Chen.
Trends in Plant Science (2024)
Highlights: Reactive oxygen species (ROS) are considered toxic substances causing oxidative damage in plants, but they are also important signaling molecules crucial for many developmental processes of root and leaf. The origin of many major ROS signaling target proteins is highly conserved, predating the evolution of land plants. ROS function for the regulation of root and leaf development with the presence of phytohormones, gasotransmitters, and target proteins, particularly mitogen-activated protein kinases (MAPKs).
Abstract: "Reactive oxygen species (ROS) are the key players in regulating developmental processes of plants. Plants have evolved a large array of gene families to facilitate the ROS-regulated developmental process in roots and leaves. However, the cellular targets of ROS during plant evolutionary development are still elusive. Here, we found early evolution and large expansions of protein families such as mitogen-activated protein kinases (MAPK) in the evolutionarily important plant lineages. We review the recent advances in interactions among ROS, phytohormones, gasotransmitters, and protein kinases. We propose that these signaling molecules act in concert to maintain cellular ROS homeostasis in developmental processes of root and leaf to ensure the fine-tuning of plant growth for better adaptation to the changing climate."
Authors: Yangdan Li, Yoshiaki Kamiyama, Fuko Minegishi, Yuki Tamura, Kota Yamashita, Sotaro Katagiri, Hinano Takase, Masahiko Otani, Ryo Tojo, Gabrielle E. Rupp, Takamasa Suzuki, Naoto Kawakami, Scott C. Peck and Taishi Umezawa.
The Plant Journal (2024)
Significance Statement: ABA-induced leaf senescence is one of the important processes involved in the translocation of nutrients from one tissue to another under drought stress. We found that group C MAP kinases phosphorylate MBD10 in response to ABA, and the two proteins constitute a regulatory pathway for ABA-induced leaf senescence in Arabidopsis.
Abstract: "Abscisic acid (ABA) is a phytohormone that promotes leaf senescence in response to environmental stress. We previously identified methyl CpG-binding domain 10 (MBD10) as a phosphoprotein that becomes differentially phosphorylated after ABA treatment in Arabidopsis. ABA-induced leaf senescence was delayed in mbd10 knockout plants but accelerated in MBD10-overexpressing plants, suggesting that MBD10 positively regulates ABA-induced leaf senescence. ABA-induced phosphorylation of MBD10 occurs in planta on Thr-89, and our results demonstrated that Thr-89 phosphorylation is essential for MBD10's function in leaf senescence. The in vivo phosphorylation of Thr-89 in MBD10 was significantly downregulated in a quadruple mutant of group C MAPKs (mpk1/2/7/14), and group C MAPKs directly phosphorylated MBD10 in vitro. Furthermore, mpk1/2/7/14 showed a similar phenotype as seen in mbd10 for ABA-induced leaf senescence, suggesting that group C MAPKs are the cognate kinases of MBD10 for Thr-89. Because group C MAPKs have been reported to function downstream of SnRK2s, our results indicate that group C MAPKs and MBD10 constitute a regulatory pathway for ABA-induced leaf senescence."
Authors: Guan-Zhu Wang, Xue Wu and Ge-Fei Hao.
Advanced Agrochem (2024)
Highlights: •Emphasize a novel mechanisms of extracellular auxin perception—ABLs and TMKs are co-receptors for extracellular auxin. •Highlight a major breakthrough in the field of auxin by Yang, Xu and their colleagues.
Abstract: "Auxin is an important phytohormone that regulates a string of vital rapid responses, and its signaling perception mechanism has been one of the hot spots of research. It has been shown that the ABP1/TMKs module is involved in regulating extracellular auxin signaling, however, the role of ABP1 as an auxin receptor is highly controversial. Therefore, the mechanism of quintessential TMKs sense extracellular auxin remains unresolved. Recently, a study identified two new auxin-binding proteins, ABL1 and ABL2, which directly interact with TMKs to perceive apoplast auxin. This groundbreaking research unravels the mystery surrounding how plants perceive extracellular auxin signals."
Authors: Yi Zhang, Yingying Xing, Xinyu Tian, Liuhui Yang, Likai Wang, Zhiyong Guan, Jiafu Jiang, Fadi Chen and Sumei Chen.
Postharvest Biology and Technology (2024)
Highlights: • Sucrose antagonizes SL-induced leaf senescence in chrysanthemum. • Various genes antagonistically regulated by SL and sucrose were revealed by RNA-seq. • CmWRKY6-Like positively regulates leaf senescence in chrysanthemum. • CmWRKY6-Like involves the antagonistic regulation of leaf senescence by SL and sugar.
Abstract: "Strigolactone (SL), a novel plant hormone, plays a vital role in promoting leaf senescence. Sugar, a nutrient source widely used to retain freshness of cut flowers, was recently reported to alleviate SL-induced leaf senescence; however, the underlying molecular mechanisms remain unclear. Leaf senescence during transportation and storage of cut chrysanthemum considerably affects its shelf life and ornamental value. In this study, we found that sucrose antagonizes SL-induced leaf senescence in chrysanthemum. Transcriptional reprogramming analysis revealed a number of differentially expressed genes antagonistically regulated by SL and sucrose, mainly including those related to SL signaling, sugar signaling, ethylene, auxin, jasmonic acid, reactive oxygen species, chlorophyll metabolism pathways, and transcription factors (such as WRKY, NAC, AP2/ERF, and MYB). Virus-based transient silencing of CmWRKY6-Like in chrysanthemum revealed that CmWRKY6-Like positively regulates leaf senescence and involves the antagonistic regulation of leaf senescence by SL and sucrose. This study provides a basis for understanding the molecular mechanisms underlying the antagonistic roles of SL- and sugar-mediated leaf senescence in chrysanthemum."
Authors: Xi Luo, Lei Guo, Ethan Tagliere, Zhenbiao Yang and Zhongchi Liu.
Current Biology (2024)
Editor's view: Using the wild strawberry as a model, Luo et al. illustrate how two transcription factors, SIMPLE LEAF1 and SALAD, regulate leaf complexity and leaf margin serration, respectively, through modulating CUC2a expression in distinct temporal and spatial domains of leaf primordia.
Highlights • Leaf serration is regulated by SALAD, a single-Myb domain protein in strawberry • CUC2a regulates leaf complexity and serration at different stages of leaf primordia • SIMPLE LEAF1 (SL1) and SALAD regulate CUC2a independently at leaf primordia • Knockouts of both SALAD homologs in Arabidopsis also lead to deeper leaf serration
Abstract: "The remarkable diversity of leaf forms allows plants to adapt to their living environment. In general, leaf diversity is shaped by leaf complexity (compound or simple) and leaf margin pattern (entire, serrated, or lobed). Prior studies in multiple species have uncovered a conserved module of CUC2-auxin that regulates both leaf complexity and margin serration. How this module is regulated in different species to contribute to the species-specific leaf form is unclear. Furthermore, the mechanistic connection between leaf complexity and leaf serration regulation is not well studied. Strawberry has trifoliate compound leaves with serrations at the margin. In the wild strawberry Fragaria vesca, a mutant named salad was isolated that showed deeper leaf serrations but normal leaf complexity. SALAD encodes a single-Myb domain protein and is expressed at the leaf margin. Genetic analysis showed that cuc2a is epistatic to salad, indicating that SALAD normally limits leaf serration depth by repressing CUC2a expression. When both Arabidopsis homologs of SALAD were knocked out, deeper serrations were observed in Arabidopsis rosette leaves, supporting a conserved function of SALAD in leaf serration regulation. We incorporated the analysis of a third strawberry mutant simple leaf 1 (sl1) with reduced leaf complexity but normal leaf serration. We showed that SL1 and SALAD independently regulate CUC2a at different stages of leaf development to, respectively, regulate leaf complexity and leaf serration. Our results provide a clear and simple mechanism of how leaf complexity and leaf serration are coordinately as well as independently regulated to achieve diverse leaf forms."
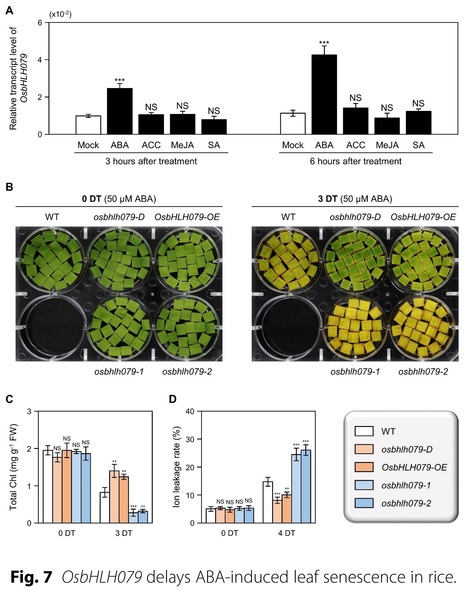
Authors: Suk-Hwan Kim, Jungwon Yoon, Hanna Kim, Sang-Ji Lee & Nam-Chon Paek.
Rice (2023)
Abstract: "Leaf senescence represents the final phase of leaf development and is characterized by a highly organized degenerative process involving the active translocation of nutrients from senescing leaves to growing tissues or storage organs. To date, a large number of senescence-associated transcription factors (sen-TFs) have been identified that regulate the initiation and progression of leaf senescence. Many of these TFs, including NAC (NAM/ATAF1/2/CUC2), WRKY, and MYB TFs, have been implicated in modulating the expression of downstream senescence-associated genes (SAGs) and chlorophyll degradation genes (CDGs) under the control of phytohormones. However, the involvement of basic helix-loop-helix (bHLH) TFs in leaf senescence has been less investigated. Here, we show that OsbHLH079 delays both natural senescence and dark-induced senescence: Overexpression of OsbHLH079 led to a stay-green phenotype, whereas osbhlh079 knockout mutation displayed accelerated leaf senescence. Similar to other sen-TFs, OsbHLH079 showed a gradual escalation in expression as leaves underwent senescence. During this process, the mRNA levels of SAGs and CDGs remained relatively low in OsbHLH079 overexpressors, but increased sharply in osbhlh079 mutants, suggesting that OsbHLH079 negatively regulates the transcription of SAGs and CDGs under senescence conditions. Additionally, we found that OsbHLH079 delays ABA-induced senescence. Subsequent RT-qPCR and dual-luciferase reporter assays revealed that OsbHLH079 downregulates the expression of ABA signaling genes, such as OsABF2, OsABF4, OsABI5, and OsNAP. Taken together, these results demonstrate that OsbHLH079 functions in delaying leaf yellowing by attenuating the ABA responses."
Authors: Haiyan Zhao, Shuyuan Wan, Yanni Huang, Xiaoqiang Li, Tiantian Jiao, Zhijun Zhang, Baiquan Ma, Lingcheng Zhu, Fengwang Ma and Mingjun Li.
The Plant Cell (2024)
Abstract: "Auxin plays important roles throughout plant growth and development. However, the mechanisms of auxin regulation of plant structure are poorly understood. In this study, we identified a transcription factor of the BARLEY B RECOMBINANT/BASIC PENTACYSTEINE (BBR/BPC) family in apple (Malus × domestica), MdBPC2. It was highly expressed in dwarf rootstocks and it negatively regulated auxin biosynthesis. Overexpression of MdBPC2 in apple decreased plant height, altered leaf morphology, and inhibited root system development. These phenotypes were due to reduced auxin levels and were restored reversed after exogenous IAA treatment. Silencing of MdBPC2 alone had no obvious phenotypic effect, while silencing both class I and class II BPCs in apple significantly increased auxin content in plants. Biochemical analysis demonstrated that MdBPC2 directly bound to the GAGA-rich element in the promoters of the auxin synthesis genes MdYUC2a and MdYUC6b, inhibiting their transcription and reducing auxin accumulation in MdBPC2 overexpression lines. Further studies established that MdBPC2 interacted with the polycomb group (PcG) protein LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) to inhibit MdYUC2a and MdYUC6b expression via methylation of histone 3 lysine 27 (H3K27me3). Silencing MdLHP1 reversed the negative effect of MdBPC2 on auxin accumulation. Our results reveal a dwarfing mechanism in perennial woody plants involving control of auxin biosynthesis by a BPC transcription factor, suggesting its use for genetic improvement of apple rootstock."
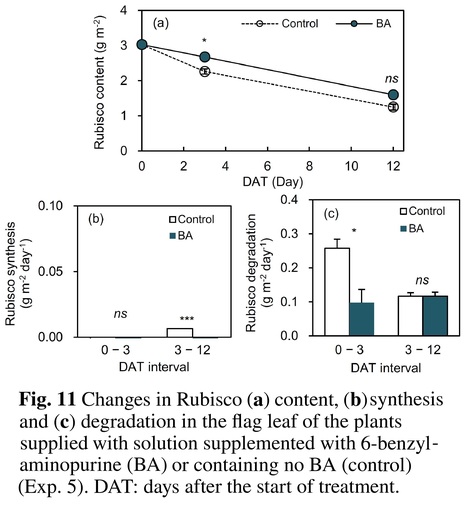
Authors: Koji Nakanishi, Hiroko Fujiki, Koichi Ozaki, Satoko Yanahara, Naoko Takeuchi, Yuji Suzuki, Tamiji Sugiyama, Amane Makino, Taiichiro Ookawa and Tadashi Hirasawa.
Plant and Soil (2023)
Abstract: "Background and Aims - Leaf senescence is accelerated by soil moisture stress during reproductive growth in rainfed paddy rice under drought and in irrigated paddy rice under intermittent irrigation for saving water or mitigating methane emissions. Leaf senescence decreases leaf photosynthetic rate (An) and grain yield. We aimed to elucidate the mechanisms underlying the An decrease under soil moisture stress. Methods - An, leaf content of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), synthesis and degradation of Rubisco and cytokinin flux from roots were compared between plants grown in moisture-deficient soil (DR-plants) and flooded or wet soil (FL- or WE-plants, respectively) during senescence in pot-grown rice (Oryza sativa L.). Results - The decreases in An and Rubisco content were larger in the DR-plants than in the FL-plants. An was closely correlated with Rubisco content during moisture stress treatment. The larger decrease of Rubisco content in the DR-plants was from increased Rubisco degradation rather than decreased synthesis. The amount of cytokinins transported from roots to shoots was smaller in the DR-plants. The application of 6-benzylaminopurine to leaves of the DR-plants suppressed Rubisco degradation. In a wilty mutant with impaired leaf hydraulic conductance, leaf senescence was significantly higher in the DR-plants than in the FL-plants, although leaf water potential of both groups decreased similarly under sunny conditions. Conclusion - The main cause of an An decrease with senescence in rice under soil moisture stress was the decrease of cytokinin flux from roots to shoots and enhanced Rubisco degradation."
|
Authors: Juan He, Xiaoyi Li, Qin Yu, Lu Peng, Li Chen, Jiajia Liu, Jianmei Wang, Xufeng Li and Yi Yang.
Plant, Cell & Environment (2024)
Summary statement: The AGC VIII kinase D6 PROTEIN KINASE (D6PK) phosphorylates PIN-FORMED (PIN) auxin efflux carriers, which is essential for auxin transport in plant cells. This study uncovers a new mechanism for D6PK stability through Cytosolic ABA Receptor Kinases-mediated phosphorylation to regulate auxin-controlled Arabidopsis growth and development.
Abstract: "The polar auxin transport is required for proper plant growth and development. D6 PROTEIN KINASE (D6PK) is required for the phosphorylation of PIN-FORMED (PIN) auxin efflux carriers to regulate auxin transport, while the regulation of D6PK stabilization is still poorly understood. Here, we found that Cytosolic ABA Receptor Kinases (CARKs) redundantly interact with D6PK, and the interactions are dependent on CARKs' kinase activities. Similarly, CARK3 also could interact with paralogs of D6PK, including D6PKL1, D6PKL2, and D6PKL3. The genetic analysis shows that D6PK acts the downstream of CARKs to regulate Arabidopsis growth, including hypocotyl, leaf area, vein formation, and the length of silique. Loss-of-function of CARK3 in overexpressing GFP-D6PK plants leads to reduce the level of D6PK protein, thereby rescues plant growth. In addition, the cell-free degradation assays indicate that D6PK is degraded through 26 S proteasome pathway, while the phosphorylation by CARK3 represses this process in cells. In summary, D6PK stabilization by the CARK family is required for auxin-mediated plant growth and development."
Authors: Muhmmad Asad Ullah Asad, Zhang Yan, Lujian Zhou, Xianyue Guan and Fangmin Cheng.
Plant Physiology and Biochemistry (2024)
Highlights: • Sugars play an essential role in the regulations of leaf senescence. • Abiotic stresses trigger sugar signaling by inducing reactive oxygen species burst. • Sugar signaling interact with plant hormones and protein kinase to regulates leaf senescence. • Abiotic stresses target sugar signaling to regulate photosynthesis inhibition and programmed cell death (PCD).
Abstract: "Plants have evolved the adaptive capacity to mitigate the negative effect of external adversities at chemical, molecular, cellular, and physiological levels. This capacity is conferred by triggering the coordinated action of internal regulatory factors, in which sugars play an essential role in the regulating chloroplast degradation and leaf senescence under various stresses. In this review, we summarize the recent findings on the senescent-associated changes in carbohydrate metabolism and its relation to chlorophyll degradation, oxidative damage, photosynthesis inhibition, programmed cell death (PCD), and sink-source relation as affected by abiotic stresses. The action of sugar signaling in regulating the initiation and progression of leaf senescence under abiotic stresses involves interactions with various plant hormones, reactive oxygen species (ROS) burst, and protein kinases. This discussion aims to elucidate the complex regulatory network and molecular mechanisms that underline sugar-induced leaf senescence in response to various abiotic stresses. The imperative role of sugar signaling in regulating plant stress responses potentially enables the production of crop plants with modified sugar metabolism. This, in turn, may facilitate the engineering of plants with improved stress responses, optimal life span and higher yield achievement."
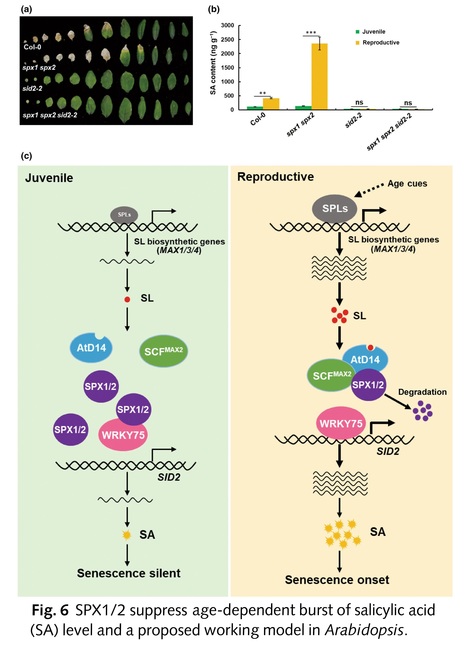
Authors: Yexing Jing, Ziyi Yang, Zongju Yang, Wanqing Bai, Ruizhen Yang, Yanjun Zhang, Kewei Zhang, Yunwei Zhang and Jiaqiang Sun.
New Phytologist (2024)
Abstract: "Leaf senescence is a complex process strictly regulated by various external and endogenous factors. However, the key signaling pathway mediating leaf senescence remains unknown. Here, we show that Arabidopsis SPX1/2 negatively regulate leaf senescence genetically downstream of the strigolactone (SL) pathway. We demonstrate that the SL receptor AtD14 and MAX2 mediate the age-dependent degradation of SPX1/2. Intriguingly, we uncover an age-dependent accumulation of SLs in leaves via transcriptional activation of SL biosynthetic genes by the transcription factors (TFs) SPL9/15. Furthermore, we reveal that SPX1/2 interact with the WRKY75 subclade TFs to inhibit their DNA-binding ability and thus repress transcriptional activation of salicylic acid (SA) biosynthetic gene SA Induction-Deficient 2, gating the age-dependent SA accumulation in leaves at the leaf senescence onset stage. Collectively, our new findings reveal a signaling pathway mediating sequential activation of SL and salicylate biosynthesis for the onset of leaf senescence in Arabidopsis."
Authors: Kangning Zhang, Hongli Xie, Jiangqi Wen, Jing Zhang, Zeng-Yu Wang, Bin Xu and Maofeng Chai.
Grass Research (2024)
Abstract: "Leaf senescence is a complex biological process regulated by development, phytohormones, and various environmental factors. For forage and turf grasses, controlling leaf senescence can greatly improve forage quality, the amenity of lawn and turf, and the grasses’ stress tolerances. Leaf senescence involves a multitude of gene regulation and metabolic changes, including the alteration of chlorophyll metabolism. Here, we summarized the recent progress of studies on leaf senescence in major forage and turf grass species, such as Medicago truncatula, M. sativa, Lolium perenne, Panicum virgatum, and Agrostis stolonifera, to provide an insight into the development of effective methods for delaying leaf senescence in grass species."
Authors: Wei Chang, Huina Zhao, Hongqiao Chen, Guixiang Jiao, Jing Yu, Bing Wang, Haiqian Xia, Boyu Meng, Xiaodong Li, Mengna Yu, Shengting Li, Mingchao Qian, Yonghai Fan, Kai Zhang, Bo Lei and Kun Lu.
Plant Physiology (2024)
Abstract: "Leaf senescence is a vital aspect of plant physiology and stress responses and is induced by endogenous factors and environmental cues. The plant-specific NAC (NAM, ATAF1/2, CUC2) transcription factor family influences growth, development, and stress responses in Arabidopsis (Arabidopsis thaliana) and other species. However, the roles of NACs in tobacco (Nicotiana tabacum) leaf senescence are still unclear. Here, we report that NtNAC56 regulates leaf senescence in tobacco. Transgenic plants overexpressing NtNAC56 (NtNAC56-OE) showed induction of senescence-related genes and exhibited early senescence and lower chlorophyll content compared to wild-type (WT) plants and the Ntnac56-19 mutant. In addition, root development and seed germination were inhibited in the NtNAC56-OE lines. Transmission electron microscopy observations accompanied by physiological and biochemical assays revealed that NtNAC56 overexpression triggers chloroplast degradation and reactive oxygen species accumulation in tobacco leaves. Transcriptome analysis demonstrated that NtNAC56 activates leaf senescence-related genes and jasmonic acid (JA) biosynthesis pathway genes. In addition, the JA content of NtNAC56-OE plants was higher than in WT plants, and JA treatment induced NtNAC56 expression. We performed DNA affinity purification sequencing to identify direct targets of NtNAC56, among which we focused on LIPOXYGENASE 5 (NtLOX5), a key gene in JA biosynthesis. A dual-luciferase reporter assay and a yeast one-hybrid assay confirmed that NtNAC56 directly binds to the TTTCTT motif in the NtLOX5 promoter. Our results reveal a mechanism whereby NtNAC56 regulates JA-induced leaf senescence in tobacco and provide a strategy for genetically manipulating leaf senescence and plant growth."
Authors: Nikita Yadav, Preeti Nagar, Abhilasha Rawat and Ananda Mustafiz.
Environmental and Experimental Botany (2024)
Highlights: • Expression of PSKR is up-regulated in the senescent leaves. • Over-expressing lines of AtPSKR1 and OsPSKR10 show an early leaf senescence phenotype. • AtPSKR1 and OsPSKR10 trigger superoxides accumulation in leaves. • AtPSKR1 and OsPSKR10 interact with the NADPH oxidases (AtRBOHD and AtRBOHF).
Abstract "Leaf senescence represents an active and regulated degeneration process. Leucine-rich repeat receptor-like protein kinase (LRR-RLK) represents a large group of cell surface receptors that play important roles in multiple biological processes, and some are also known to regulate the senescence process. In the present study, we elucidated the involvement of Phytosulfokine Receptor (PSKR) gene in the regulation of leaf senescence. The expression of PSKR1 of Arabidopsis was found to be up-regulated during leaf senescence. The over-expressing lines of AtPSKR1 and its rice homologue OsPSKR10 displayed an early leaf senescent phenotype in Arabidopsis, while the late senescence in mutant line of AtPSKR1 supported the idea that PSKR positively regulate the process of leaf senescence. Furthermore, the leaves of the overexpressing lines displayed increased superoxide accumulation compared to mutant lines, along with higher expression of NADPH oxidases (AtRBOHD and AtRBOHF). Importantly, our findings demonstrates that both AtPSKR1 and OsPSKR10 interact with AtRBOHD and AtRBOHF and thereby instigating superoxide accumulation in the leaves through NADPH oxidases."
Authors: Foziya Altaf, Shazia Parveen, Sumira Farooq, Mohammad Lateef Lone, Aehsan Ul Haq and Inayatullah Tahir
Theoretical and Experimental Plant Physiology (2024)
Abstract: "Due to the already strained and severely challenged agricultural ecosystems of the modern world, predicted changes in life cycle of plants, including leaf senescence are receiving significant attention from stakeholders. The onset, progression and terminal phases of leaf senescence are greatly influenced by plant hormones. The senescence of leaves is accelerated by ethylene, jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA), brassinosteroids and strigolactones (SLs), whereas it is postponed by cytokinins (CKs), gibberellic acid (GA) and auxins. The crosstalk and signal transduction pathways between these growth regulators have been found to regulate leaf senescence by orchestrating various developmental and environmental factors. Premature leaf senescence lessens the plant’s nutritional capacity and shortens the vegetative production schedule, prompting an early transition from the vegetative to the reproductive stage and diminishing crop potential. As a result, a complete understanding of leaf senescence and finding novel ways to delay it is crucial for agricultural productivity. The ability to manipulate leaf senescence for agricultural enhancement has been made possible by significant advances in physiological and molecular awareness of leaf senescence. Although studies pertaining to leaf senescence have been given steadily more attention, there are still numerous challenges that need to be resolved. In this perspective, this review focuses on current advances in understanding the leaf senescence by molecular and genetic analyses with an emphasis on hormonal regulation of leaf senescence. We also hypothesize future research to better comprehend leaf senescence by employing various current technologies."
Author: Enrico Scarpella.
Annual Review of Plant Biology (2024)
Abstract: "Leaves form veins whose patterns vary from a single vein running the length of the leaf to networks of staggering complexity where huge numbers of veins connect to other veins at both ends. For the longest time, vein formation was thought to be controlled only by the polar, cell-to-cell transport of the plant hormone auxin; recent evidence suggests that is not so. Instead, it turns out that vein patterning features are best accounted for by a combination of polar auxin transport, facilitated auxin diffusion through plasmodesmata intercellular channels, and auxin signal transduction—though the latter's precise contribution remains unclear. Equally unclear remain the sites of auxin production during leaf development, on which that vein patterning mechanism ought to depend. Finally, whether that vein patterning mechanism can account for the variety of vein arrangements found in nature remains unknown. Addressing those questions will be the exciting challenge of future research."
Authors: Arielle Homayouni, Suresh Damodaran, Katherine Schreiber, Marta Michniewicz, Lauren Gunther and Lucia C Strader.
bioRxiv (2024)
Abstract: "Proper spatiotemporal distribution of the phytohormone auxin throughout plant tissues mediates a variety of developmental processes. Auxin levels are tightly regulated via de novo synthesis, transport, and conversion from its conjugated forms and precursors. These levels can be regulated through conversion of the auxin precursor, indole 3-butyric acid (IBA), into the active auxin, indole-3-acetic acid (IAA), in a peroxisomal β-oxidation process. Defects in IBA-to-IAA conversion cause multiple developmental defects in Arabidopsis, demonstrating IBA-derived IAA is physiologically important to the active auxin pool. Similar to IAA, transport of IBA modulates development. However, the mechanisms governing transport of this molecule remain largely unknown. Here, we identify a mutation in the ABCC10 gene of Arabidopsis that suppresses the abcg36 hypersensitivity to IBA and its synthetic analog, 2,4-dichlorophenoxy butyric acid (2,4-DB) and the abcg36 hyperaccumulation of [3H]-IBA. We found that ABCC10 acts as a direct vacuolar transporter of IBA. Further, ABCC10 is necessary for proper development of the root apical meristem and leaf tissue. Our findings uncover a previously uncharacterized method of IBA transport that regulates aspects of plant development."
Authors: Zhuang Li, Xiangguang Lyu, Hongyu Li, Qichao Tu, Tao Zhao, Jun Liu and Bin Liu.
Nature Communications (2024)
Editor's view: This study provides insights into how shade induces leaf senescence in soybean. The reduction of blue light intensity deactivates GmCRY1s, leading to the degradation of GmRGAs and the upregulation of WRKY100, ultimately promoting leaf senescence.
Abstract: "Leaf senescence is a crucial trait that has a significant impact on crop quality and yield. Previous studies have demonstrated that light is a key factor in modulating the senescence process. However, the precise mechanism by which plants sense light and control senescence remains largely unknown, particularly in crop species. In this study, we reveal that the reduction in blue light under shading conditions can efficiently induce leaf senescence in soybean. The blue light receptors GmCRY1s rather than GmCRY2s, primarily regulate leaf senescence in response to blue light signals. Our results show that GmCRY1s interact with DELLA proteins under light-activated conditions, stabilizing them and consequently suppressing the transcription of GmWRKY100 to delay senescence. Conversely, LBL reduces the interaction between GmCRY1s and the DELLA proteins, leading to their degradation and premature senescence of leaves. Our findings suggest a GmCRY1s-GmDELLAs-GmWRKY100 regulatory cascade that is involved in mediating LBL-induced leaf senescence in soybean, providing insight into the mechanism of how light signals regulate leaf senescence. Additionally, we generate GmWRKY100 knockout soybeans that show delayed leaf senescence and improved yield under natural field conditions, indicating potential applications in enhancing soybean production by manipulating the leaf senescence trait."
Authors: María Florencia Mammarella, Leandro Lucero, Nosheen Hussain, Aitor Muñoz-Lopez, Ying Huang, Lucia Ferrero, Guadalupe L Fernandez-Milmanda, Pablo Manavella, Moussa Benhamed, Martin Crespi, Carlos L. Ballare, José Gutiérrez Marcos, Pilar Cubas and Federico Ariel.
EMBO Journal (2023)
Synopsis: The Arabidopsis lncRNA APOLO influences chromatin architecture to regulate transcription of specific genes, including auxin-responsive genes which are involved in the plant's response to shade avoidance. Here, APOLO is shown to respond to changes in light by influencing transcription and plant behavior. APOLO modulates the expression of BRANCHED1 (BRC1), a master regulator of shoot branching in Arabidopsis, through chromatin looping around the BRC1 promoter. APOLO-mediated chromatin looping is influenced by changes in light exposure and regulates branching behavior. Low-red/far-red light ratio-mediated leaf hyponasty depends on APOLO regulation of auxin-related genes. In vitro-transcribed APOLO directly sprayed onto plants is sufficient to alter plant auxin homeostasis in response to light conditions.
Abstract: "The long noncoding RNA (lncRNA) AUXIN-REGULATED PROMOTER LOOP (APOLO) recognizes a subset of target loci across the Arabidopsis thaliana genome by forming RNA–DNA hybrids (R-loops) and modulating local three-dimensional chromatin conformation. Here, we show that APOLO regulates shade avoidance syndrome by dynamically modulating expression of key factors. In response to far-red (FR) light, expression of APOLO anti-correlates with that of its target BRANCHED1 (BRC1), a master regulator of shoot branching in Arabidopsis thaliana. APOLO deregulation results in BRC1 transcriptional repression and an increase in the number of branches. Accumulation of APOLO transcription fine-tunes the formation of a repressive chromatin loop encompassing the BRC1 promoter, which normally occurs only in leaves and in a late response to far-red light treatment in axillary buds. In addition, our data reveal that APOLO participates in leaf hyponasty, in agreement with its previously reported role in the control of auxin homeostasis through direct modulation of auxin synthesis gene YUCCA2, and auxin efflux genes PID and WAG2. We show that direct application of APOLO RNA to leaves results in a rapid increase in auxin signaling that is associated with changes in the plant response to far-red light. Collectively, our data support the view that lncRNAs coordinate shade avoidance syndrome in A. thaliana, and reveal their potential as exogenous bioactive molecules. Deploying exogenous RNAs that modulate plant–environment interactions may therefore become a new tool for sustainable agriculture."
Authors: Andrey A. Kotov and Liudmila M. Kotova.
Journal of Experimental Botany (2023)
Abstract: "Cammarata et al. (2023) show that in the model non-vascular plant Physcomitrium patens the growth hormone indole acetic acid (IAA) can inhibit growth and development of leaves (phyllids) from shoots, while cytokinin (CK) eliminates this effect. Most significantly, the effect on growth of the observed antagonism between IAA and CK has also been reported for vascular plants in the literature. These new data from moss emphasize the evolutionary preservation of this interaction between these hormones in the control of shoot growth."
|


 Your new post is loading...
Your new post is loading...