Your new post is loading...
 Your new post is loading...
Authors: Xiaolin Liu, Shusong Zheng, Shuiquan Tian, Yaoqi Si, Shengwei Ma, Hong-Qing Ling and Jianqing Niu.
Theoretical and Applied Genetics (2024)
Key message: Discovery of Rht27, a dwarf gene in wheat, showed potential in enhancing grain yield by reducing plant height.
Abstract: "Plant height plays a crucial role in crop architecture and grain yield, and semi-dwarf Reduced Height (Rht) alleles contribute to lodging resistance and were important in “Green Revolution.” However, the use of these alleles is associated with some negative side effects in some environments, such as reduced coleoptile length, low nitrogen use efficiency, and reduced yield. Therefore, novel dwarf gene resources are needed to pave an alternative route to overcome these side effects. In this study, a super-dwarf mutant rht27 was obtained by the mutagenesis of G1812 (Triticum urartu, the progenitor of the A sub-genome of common wheat). Genetic analysis revealed that the dwarf phenotype was regulated by a single recessive genetic factor. The candidate region for Rht27 was narrowed to a 1.55 Mb region on chromosome 3, within which we found two potential candidate genes that showed polymorphisms between the mutant and non-mutagenized G1812. Furthermore, the natural variants and elite haplotypes of the two candidates were investigated in a natural population of common wheat. The results showed that the natural variants affect grain yield components, and the dwarf haplotypes show the potential in improving agronomic traits and grain yield. Although the mutation in Rht27 results in severe dwarf phenotype in T. urartu, the natural variants in common wheat showed desirable phenotype, which suggests that Rht27 has the potential to improve wheat yield by utilizing its weak allelic mutation or fine-tuning its expression level."
Authors: Amanpreet Kaur, Norman B. Best, Thomas Hartwig, Josh Budka, Rajdeep Khangura, Steven McKenzie, Alejandro Aragón-Raygoza, Josh Strable, Burkhard Schulz and Brian P. Dilkes.
Plant Physiology (2023)
One-sentence summary: Molecular identity of a maize semidwarf mutant reveals a role for the maize GRAS domain transcription factor ortholog of DWARF AND LOW TILLERING in brassinosteroid signaling.
Abstract: "Brassinosteroids (BR) and gibberellins (GA) regulate plant height and leaf angle in maize (Zea mays). Mutants with defects in BR or GA biosynthesis or signaling identify components of these pathways and enhance our knowledge about plant growth and development. In this study, we characterized three recessive mutant alleles of GRAS transcription factor 42 (gras42) in maize, a GRAS transcription factor gene orthologous to the DWARF AND LOW TILLERING (DLT) gene of rice (Oryza sativa). These maize mutants exhibited semi-dwarf stature, shorter and wider leaves, and more upright leaf angle. Transcriptome analysis revealed a role for GRAS42 as a determinant of BR signaling. Analysis of the expression consequences from loss of GRAS42 in the gras42-mu1021149 mutant indicated a weak loss of BR signaling in the mutant, consistent with its previously demonstrated role in BR signaling in rice. Loss of BR signaling was also evident by the enhancement of weak BR biosynthetic mutant alleles in double mutants of nana plant1-1 and gras42-mu1021149. The gras42-mu1021149 mutant had little effect on GA-regulated gene expression, suggesting that GRAS42 is not a regulator of core GA signaling genes in maize. Single cell expression data identified gras42 expressed among cells in the G2/M phase of the cell cycle consistent with its previously demonstrated role in cell cycle gene expression in Arabidopsis (Arabidopsis thaliana). Cis-acting natural variation controlling GRAS42 transcript accumulation was identified by expression genome-wide association study (eGWAS) in maize. Our results demonstrate a conserved role for GRAS42/SCARECROW-LIKE 28 (SCL28)/DLT in BR signaling, clarify the role of this gene in GA signaling, and suggest mechanisms of tillering and leaf angle control by BR."
Authors: Xumin Xiang, Hongli Yang, Xi Yuan, Xue Dong, Sihua Mai, Qianqian Zhang, Limiao Chen, Dong Cao, Haifeng Chen, Wei Guo and Li Li.
Plant Cell Reports (2024)
Key message: The study on the GmDWF1-deficient mutant dwf1 showed that GmDWF1 plays a crucial role in determining soybean plant height and yield by influencing the biosynthesis of brassinosteroids.
Abstract: "Soybean has not adopted the Green Revolution, such as reduced height for increased planting density, which have proven beneficial for cereal crops. Our research identified the soybean genes GmDWF1a and GmDWF1b, homologous to Arabidopsis AtDWF1, and found that they are widely expressed, especially in leaves, and linked to the cellular transport system, predominantly within the endoplasmic reticulum and intracellular vesicles. These genes are essential for the synthesis of brassinosteroids (BR). Single mutants of GmDWF1a and GmDWF1b, as well as double mutants of both genes generated through CRISPR/Cas9 genome editing, exhibit a dwarf phenotype. The single-gene mutant exhibits moderate dwarfism, while the double mutant shows more pronounced dwarfism. Despite the reduced stature, all types of mutants preserve their node count. Notably, field tests have shown that the single GmDWF1a mutant produced significantly more pods than wild-type plants. Spraying exogenous brassinolide (BL) can compensate for the loss in plant height induced by the decrease in endogenous BRs. Comparing transcriptome analyses of the GmDWF1a mutant and wild-type plants revealed a significant impact on the expression of many genes that influence soybean growth. Identifying the GmDWF1a and GmDWF1b genes could aid in the development of compact, densely planted soybean varieties, potentially boosting productivity."
Authors: Ting Li, Yongqin Wang, Annelore Natran, Yi Zhang, Hao Wang, Kangxi Du, Peng Qin, Hua Yuan, Weilan Chen, Bin Tu, Dirk Inzé and Marieke Dubois.
New Phytologist (2024)
Abstract: "Gibberellic acid (GA) plays a central role in many plant developmental processes and is crucial for crop improvement. DELLA proteins, the core suppressors in the GA signaling pathway, are degraded by GA via the 26S proteasomal pathway to release the GA response. However, little is known about the phosphorylation-mediated regulation of DELLA proteins. In this study, we combined GA response assays with protein–protein interaction analysis to infer the connection between Arabidopsis thaliana DELLAs and the C-TERMINAL DOMAIN PHOSPHATASE-LIKE 3 (CPL3), a phosphatase involved in the dephosphorylation of RNA polymerase II. We show that CPL3 directly interacts with DELLA proteins and promotes DELLA protein stability by inhibiting its degradation by the 26S proteasome. Consequently, CPL3 negatively modulates multiple GA-mediated processes of plant development, including hypocotyl elongation, flowering time, and anthocyanin accumulation. Taken together, our findings demonstrate that CPL3 serves as a novel regulator that could improve DELLA stability and thereby participate in GA signaling transduction."
Authors: Barbora Ndreca, Alison Huttly, Sajida Bibi, Carlos Bayon, George Lund, Joshua Ham, Rocío Alarcón-Reverte, John Addy, Danuše Tarkowská, Stephen Pearce, Peter Hedden, Stephen G. Thomas and Andrew L. Phillips.
Research Square (2024)
Abstract: "Background - Semi-dwarfing alleles are used widely in cereals to confer improved lodging resistance and assimilate partitioning. The most widely deployed semi-dwarfing alleles in rice and barley encode the gibberellin (GA)-biosynthetic enzyme GA 20-OXIDASE2 (GA20OX2). The hexaploid wheat genome carries three homoeologous copies of GA20OX2, and because of functional redundancy, loss-of-function alleles of a single homoeologue would not be selected in wheat breeding programmes. Instead, approximately 70% of wheat cultivars carry gain-of-function mutations in REDUCED HEIGHT 1 (RHT1) genes that encode negative growth regulators and are degraded in response to GA. Semi-dwarf Rht-B1b or Rht-D1b alleles encode proteins that are insensitive to GA-mediated degradation. However, because RHT1 is expressed ubiquitously these alleles have pleiotropic effects that confer undesirable traits in some environments. Results - We have applied reverse genetics to combine loss-of-function alleles in all three homoeologues of wheat GA20OX2 and its paralogue GA20OX1 and evaluated their performance in three years of field trials. ga20ox1 mutants exhibited a mild height reduction (approximately 3%) suggesting GA20OX1 plays a minor role in stem elongation in wheat. ga20ox2 mutants have reduced GA1 content and are 12-32% shorter than their wild-type segregants, comparable to the effect of the Rht-D1b ‘Green Revolution’ allele. The ga20ox2 mutants showed no significant negative effects on yield components, although these alleles should be evaluated in different genetic backgrounds and environments. Conclusions - Our study demonstrates that induced mutagenesis can expand genetic variation in polyploid crops to uncover novel alleles and that mutations in GA20OX2 could have utility in wheat breeding as alternative semi-dwarfing alleles.
Authors: Jiahong Lv, Yi Feng, Longmei Zhai, Lizhong Jiang, Yue Wu, Yimei Huang, Runqi Yu, Ting Wu, Xinzhong Zhang, Yi Wang and Zhenhai Han.
Horticulture Research (2024)
Abstract: "Apple rootstock dwarfing and dense planting are common practices in apple farming. However, the dwarfing mechanisms are not understood. In this study, the expression of MdARF3 in the root system of dwarfing rootstock 'M9' was lower than in the vigorous rootstock from Malus micromalus due to the deletion of the WUSATAg element in the promoter of the 'M9' genotype. Notably, this deletion variation was significantly associated with dwarfing rootstocks. Subsequently, transgenic tobacco (Nicotiana tabacum) cv. Xanthi was generated with the ARF3 promoter from 'M9' and M. micromalus genotypes. The transgenic apple with 35S:: MdARF3 was also obtained. The transgenic tobacco and apple with the highly expressed ARF3 had a longer root system and a higher plant height phenotype. Furthermore, the yeast one-hybrid, luciferase, electrophoretic mobility shift assays, and Chip-qPCR identified MdWOX4-1 in apples that interacted with the pMm-ARF3 promoter but not the pM9-ARF3 promoter. Notably, MdWOX4-1 significantly increased the transcriptional activity of MdARF3 and MdLBD16-2. However, MdARF3 significantly decreased the transcriptional activity of MdLBD16-2. Further analysis revealed that MdARF3 and MdLBD16-2 were temporally expressed during different stages of lateral root development. pMdLBD16-2 was mainly expressed during the early stage of lateral root development, which promoted lateral root production. On the contrary, pMmARF3 was expressed during the late stage of lateral root development to promote elongation. The findings in this study will shed light on the genetic causes of apple plant dwarfism and provide strategies for molecular breeding of dwarfing apple rootstocks."
Authors: Xiu Fang, Hao Wu, Wanchang Huang, Zhongxian Ma, Yue Jia, Yongwei Min, Qing Ma and Ronghao Cai.
Plant Physiology and Biochemistry (2024)
Highlights: • Mutation of the ZmWRKY92 gene resulted in decreased plant height in maize. • ZmWRKY92 is a nuclear localized protein with transactivation activity in yeast. • ZmWRKY92 binds to the W-box element in the promoter of GA synthesis-related genes.
Abstract: "The plant height is a crucial agronomic trait in contemporary maize breeding. Appropriate plant height can improve crop lodging resistance, increase the planting density and harvest index of crops, and thus contribute to stable and increased yields. In this study, molecular characterization showed that ZmWRKY92 is a nuclear protein and has transcriptional activation in yeast. ZmWRKY92 can specifically bind to the W-box (TTGACC), which was confirmed by double LUC experiments and Yeast one-hybrid assays. Subsequently we screened wrky92 mutants from a library of ethyl methanesulfonate (EMS)-induced mutants. The mutation of a base in ZmWRKY92 leading to the formation of a truncated protein variant is responsible for the dwarfing phenotype of the mutant, which was further verified by allelic testing. Detailed phenotypic analysis revealed that wrky92 mutants have shorter internodes due to reduced internode cell size and lower levels of GA3 and IAA. Transcriptome analysis revealed that the ZmWRKY92 mutation caused significant changes in the expression of genes related to plant height in maize. Additionally, ZmWRKY92 was found to interact with the promoters of ZmGA20ox7 and ZmGID1L2, which are associated with GA synthesis. This study shows that ZmWRKY92 significantly affects the plants height in maize and is crucial in identifying new varieties suitable for growing in high-density conditions."
Authors: Haiqiang Zhang, Zichen Liu, Yunxiao Wang, Siyu Mu, Hongzhong Yue, Yanjie Luo, Zhengao Zhang, Yuhong Li and Peng Chen.
Theoretical and Applied Genetics (2024)
Key message A novel super compact mutant, scp-3, was identified using map-based cloning in cucumber. The CsDWF7 gene encoding a delta7 sterol C-5(6) desaturase was the candidate gene of scp-3.
Abstract: "Mining dwarf genes is important in understanding stem growth in crops. However, only a small number of dwarf genes have been cloned or characterized. Here, we characterized a cucumber (Cucumis sativus L.) dwarf mutant, super compact 3 (scp-3), which displays shortened internodes and dark green leaves with a wrinkled appearance. The photosynthetic rate of scp-3 is significantly lower than that of the wild type. The dwarf phenotype of scp-3 mutant can be partially rescued by the exogenous brassinolide (BL) application, and the endogenous brassinosteroids (BRs) levels in the scp-3 mutant were significantly lower compared to the wild type. Microscopic examination revealed that the reduced internode length in scp-3 resulted from a decrease in cell size. Genetic analysis showed that the dwarf phenotype of scp-3 was controlled by a single recessive gene. Combined with bulked segregant analysis and map-based cloning strategy, we delimited scp-3 locus into an 82.5 kb region harboring five putative genes, but only one non-synonymous mutation (A to T) was discovered between the mutant and its wild type in this region. This mutation occurred within the second exon of the CsGy4G017510 gene, leading to an amino acid alteration from Leu156 to His156. This gene encodes the CsDWF7 protein, an analog of the Arabidopsis DWF7 protein, which is known to be involved in the biosynthesis of BRs. The CsDWF7 protein was targeted to the cell membrane. In comparison to the wild type, scp-3 exhibited reduced CsDWF7 expression in different tissues. These findings imply that CsDWF7 is essential for both BR biosynthesis as well as growth and development of cucumber plants."
Authors: Guangyan Yang, Manyi Sun, Lester Brewer, Zikai Tang, Niels Nieuwenhuizen, Janine Cooney, Shaozhuo Xu, Jiawen Sheng, Christelle Andre, Cheng Xue, Ria Rebstock, Bo Yang, Wenjing Chang, Yueyuan Liu, Jiaming Li, Runze Wang, Mengfan Qin, Cyril Brendolise, Andrew C. Allan, Richard V. Espley, Kui Lin-Wang and Jun Wu.
Plant Biotechnology Journal (2024)
Abstract: "Variation in anthocyanin biosynthesis in pear fruit provides genetic germplasm resources for breeding, while dwarfing is an important agronomic trait, which is beneficial to reduce the management costs and allow for the implementation of high-density cultivation. Here, we combined bulked segregant analysis (BSA), quantitative trait loci (QTL), and structural variation (SV) analysis to identify a 14-bp deletion which caused a frame shift mutation and resulted in the premature translation termination of a B-box (BBX) family of zinc transcription factor, PyBBX24, and its allelic variation termed PyBBX24ΔN14. PyBBX24ΔN14 overexpression promotes anthocyanin biosynthesis in pear, strawberry, Arabidopsis, tobacco, and tomato, while that of PyBBX24 did not. PyBBX24ΔN14 directly activates the transcription of PyUFGT and PyMYB10 through interaction with PyHY5. Moreover, stable overexpression of PyBBX24ΔN14 exhibits a dwarfing phenotype in Arabidopsis, tobacco, and tomato plants. PyBBX24ΔN14 can activate the expression of PyGA2ox8 via directly binding to its promoter, thereby deactivating bioactive GAs and reducing the plant height. However, the nuclear localization signal (NLS) and Valine-Proline (VP) motifs in the C-terminus of PyBBX24 reverse these effects. Interestingly, mutations leading to premature termination of PyBBX24 were also identified in red sports of un-related European pear varieties. We conclude that mutations in PyBBX24 gene link both an increase in pigmentation and a decrease in plant height."
Authors: Fereshteh Jafari, Baobao Wang, Haiyang Wang and Junjie Zou.
Journal of Integrative Plant Biology (2023)
Abstract: "Maize is a major staple crop widely used as food, animal feed, and raw materials in industrial production. High-density planting is a major factor contributing to the continuous increase of maize yield. However, high planting density usually triggers a shade avoidance response and causes increased plant height and ear height, resulting in lodging and yield loss. Reduced plant height and ear height, more erect leaf angle, reduced tassel branch number, earlier flowering, and strong root system architecture are five key morphological traits required for maize adaption to high-density planting. In this review, we summarize recent advances in deciphering the genetic and molecular mechanisms of maize involved in response to high-density planting. We also discuss some strategies for breeding advanced maize cultivars with superior performance under high-density planting conditions."
Authors: Xiujie Liu, Kai Huang and Chengcai Chu.
Plant Communications (2024)
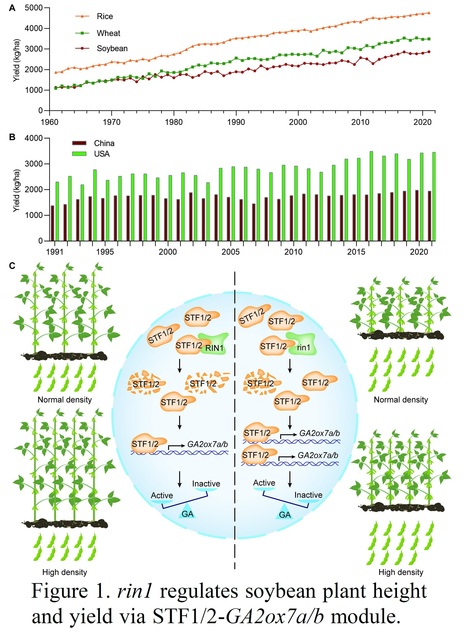
Excerpts: "Very recently, Li et al. (2023) identified an ideal allele for compact plant architecture in soybean, reduced internode 1 (rin1), that downregulates plant height by reducing internode length and increases total yield per plot under dense planting conditions (Figure 1C). Li et al. have screened out a dwarf mutant from the mutant library in the background of Heinong 35 (HN35), an elite soybean cultivar in China. Interestingly, the reduced plant height of the mutant was mainly caused by the shortened internode, and the yield per plant of rin1 is higher than HN35 (Li et al., 2023). With F2 segregating population derived from the cross between rin1 and Heihe 43 (HH43), another elite cultivar, as well as a residual heterozygous inbred population from the F2 segregating population, the candidate gene of rin1 was cloned."
"In summary, the work of Li et al. (2023) that not only provides a potential elite allele, rin1, for soybean GR, but also uncovers the molecular mechanism of plant height and internode length regulation by RIN1, making a groundbreaking advance in soybean breeding for dense planting to enhance grain yield. Introducing rin1 into more elite soybean cultivars would be an effective strategy to promote the leap of soybean production, especially for China, which has a substantially lower soybean yield compared to USA, which has benefited from soybean varieties with enhanced lodging resistance to adapt to high-density planting (Figure 1B)."
Authors: Xueya Zhao, Kunpeng Zhang, Huidong Zhang, Mengxi Bi, Yi He, Yiqing Cui, Changhua Tan, Jian Ma and Mingfang Qi.
Frontiers in Plant Science (2023)
Abstract: "Plant height is an important agronomic trait. Dwarf varieties present several advantages, such as lodging resistance, increased yield, and suitability for mechanized harvesting, which are crucial for crop improvement. However, limited research is available on dwarf tomato varieties suitable for production. In this study, we report a novel short internode mutant named “short internode and pedicel (sip)” in tomato, which exhibits marked internode and pedicel shortening due to suppressed cell elongation. This mutant plant has a compact plant structure and compact inflorescence, and has been demonstrated to produce more fruits, resulting in a higher harvest index. Genetic analysis revealed that this phenotype is controlled by a single recessive gene, SlSIP. BSA analysis and KASP genotyping indicated that ERECTA (ER) is the possible candidate gene for SlSIP, which encodes a leucine-rich receptor-like kinase. Additionally, we obtained an ER functional loss mutant using the CRISPR/Cas9 gene-editing technology. The 401st base A of ER is substituted with T in sip, resulting in a change in the 134th amino acid from asparagine (N) to isoleucine (I). Molecular dynamics(MD) simulations showed that this mutation site is located in the extracellular LRR domain and alters nearby ionic bonds, leading to a change in the spatial structure of this site. Transcriptome analysis indicated that the genes that were differentially expressed between sip and wild-type (WT) plants were enriched in the gibberellin metabolic pathway. We found that GA3 and GA4 decreased in the sip mutant, and exogenous GA3 restored the sip to the height of the WT plant. These findings reveal that SlSIP in tomatoes regulates stem elongation by regulating gibberellin metabolism. These results provide new insights into the mechanisms of tomato dwarfing and germplasm resources for breeding dwarfing tomatoes."
Authors: Shichen Li, Zhihui Sun, Qing Sang, Chao Qin, Lingping Kong, Xin Huang, Huan Liu, Tong Su, Haiyang Li, Milan He, Chao Fang, Lingshuang Wang, Shuangrong Liu, Bin Liu, Baohui Liu, Xiangdong Fu, Fanjiang Kong and Sijia Lu
Nature Communications (2023)
Editor's view: Many cereal crops have been bred to be more compact to allow high-density planting, but soybean has remained relatively overlooked. Here, the authors describe a compact soybean mutant, reduced internode 1, that significantly enhances grain yield under high-density planting conditions compared to an elite cultivar.
Abstract: "Major cereal crops have benefitted from Green Revolution traits such as shorter and more compact plants that permit high-density planting, but soybean has remained relatively overlooked. To balance ideal soybean yield with plant height under dense planting, shortening of internodes without reducing the number of nodes and pods is desired. Here, we characterized a short-internode soybean mutant, reduced internode 1 (rin1). Partial loss of SUPPRESSOR OF PHYA 105 3a (SPA3a) underlies rin1. RIN1 physically interacts with two homologs of ELONGATED HYPOCOTYL 5 (HY5), STF1 and STF2, to promote their degradation. RIN1 regulates gibberellin metabolism to control internode development through a STF1/STF2–GA2ox7 regulatory module. In field trials, rin1 significantly enhances grain yield under high-density planting conditions comparing to its wild type of elite cultivar. rin1 mutants therefore could serve as valuable resources for improving grain yield under high-density cultivation and in soybean–maize intercropping systems. Many cereal crops have been bred to be more compact to allow high-density planting, but soybean has remained relatively overlooked. Here, the authors describe a compact soybean mutant, reduced internode 1, that significantly enhances grain yield under high-density planting conditions compared to an elite cultivar."
|
Authors: Barbora Ndreca, Alison Huttly, Sajida Bibi, Carlos Bayon, George Lund, Joshua Ham, Rocío Alarcón-Reverte, John Addy, Danuše Tarkowská, Stephen Pearce, Peter Hedden, Stephen G. Thomas and Andrew L. Phillips.
BMC Plant Biology (2024)
Abstract: Background - Semi-dwarfing alleles are used widely in cereals to confer improved lodging resistance and assimilate partitioning. The most widely deployed semi-dwarfing alleles in rice and barley encode the gibberellin (GA)-biosynthetic enzyme GA 20-OXIDASE2 (GA20OX2). The hexaploid wheat genome carries three homoeologous copies of GA20OX2, and because of functional redundancy, loss-of-function alleles of a single homoeologue would not be selected in wheat breeding programmes. Instead, approximately 70% of wheat cultivars carry gain-of-function mutations in REDUCED HEIGHT 1 (RHT1) genes that encode negative growth regulators and are degraded in response to GA. Semi-dwarf Rht-B1b or Rht-D1b alleles encode proteins that are insensitive to GA-mediated degradation. However, because RHT1 is expressed ubiquitously these alleles have pleiotropic effects that confer undesirable traits in some environments. Results - We have applied reverse genetics to combine loss-of-function alleles in all three homoeologues of wheat GA20OX2 and its paralogue GA20OX1 and evaluated their performance in three years of field trials. ga20ox1 mutants exhibited a mild height reduction (approximately 3%) suggesting GA20OX1 plays a minor role in stem elongation in wheat. ga20ox2 mutants have reduced GA1 content and are 12–32% shorter than their wild-type segregants, comparable to the effect of the Rht-D1b ‘Green Revolution’ allele. The ga20ox2 mutants showed no significant negative effects on yield components in the spring wheat variety ‘Cadenza’. Conclusions - Our study demonstrates that chemical mutagenesis can expand genetic variation in polyploid crops to uncover novel alleles despite the difficulty in identifying appropriate mutations for some target genes and the negative effects of background mutations. Field experiments demonstrate that mutations in GA20OX2 reduce height in wheat, but it will be necessary to evaluate the effect of these alleles in different genetic backgrounds and environments to determine their value in wheat breeding as alternative semi-dwarfing alleles.
Authors: Xing Wang, Zhaobin Ren, Shipeng Xie, Zhaohu Li, Yuyi Zhou and Liusheng Duan.
Plant Physiology (2024)
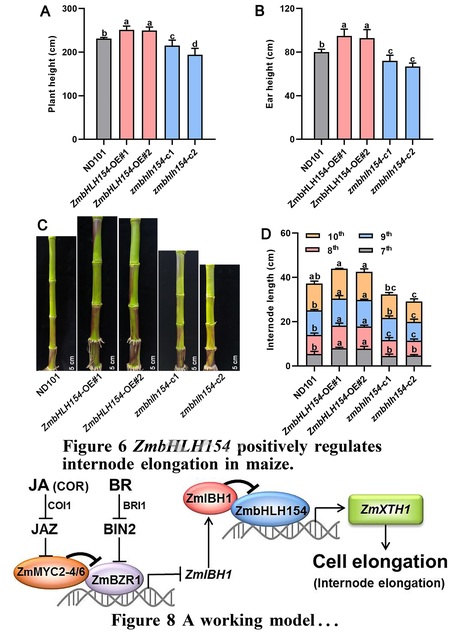
One-sentence summary: A jasmonate mimic regulates a basic helix-loop-helix network by attenuating brassinosteroid signaling, which represses expression of a cell wall–related gene and inhibits internode elongation in maize.
Abstract: "Lodging restricts growth, development, and yield formation in maize (Zea mays L.). Shorter internode length is beneficial for lodging tolerance. However, although brassinosteroids (BRs) and jasmonic acid (JA) are known to antagonistically regulate internode growth, the underlying molecular mechanism is still unclear. In this study, application of the JA mimic coronatine (COR) inhibited basal internode elongation at the jointing stage and repressed expression of the cell wall-related gene XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 1 (ZmXTH1), whose overexpression in maize plants promotes internode elongation. We demonstrated that the basic helix–loop–helix (bHLH) transcription factor ZmbHLH154 binds directly to the ZmXTH1 promoter and induces its expression, whereas the bHLH transcription factor ILI1 BINDING BHLH 1 (ZmIBH1) inhibits this transcriptional activation by forming a heterodimer with ZmbHLH154. Overexpressing ZmbHLH154 led to longer internodes, whereas zmbhlh154 mutants had shorter internodes than the wild type. The core JA-dependent transcription factors ZmMYC2-4 and ZmMYC2-6 interacted with BRASSINAZOLE RESISTANT 1 (ZmBZR1), a key factor in BR signaling, and these interactions eliminated the inhibitory effect of ZmBZR1 on its downstream gene ZmIBH1. Collectively, these results reveal a signaling module in which JA regulates a bHLH network by attenuating BR signaling to inhibit ZmXTH1 expression, thereby regulating cell elongation in maize."
Authors: Jiujun Du, Hantian Wei, Xueqin Song, Lei Zhang and Jianjun Hu.
Plant Science (2024)
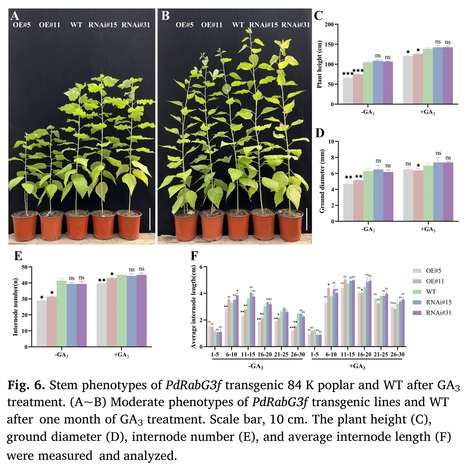
Highlights: • PdRabG3f inhibited the vertical elongation of poplar by suppressing internode elongation. • PdRabG3f inhibited xylem development by suppressing cambium division and differentiation. • PdRabG3f interfered with gibberellin-mediated internode elongation and xylem developing in poplar.
Abstract: "As a member of the small GTPases family, Rab GTPases play a key role in specifying transport pathways in the intracellular membrane trafficking system and are involved in plant growth and development. By quantitative trait locus (QTL) mapping, PdRabG3f was identified as a candidate gene associated with shoot height in a hybrid offspring of Populus deltoides ‘Danhong’ × Populus simonii ‘Tongliao1’. PdRabG3f localized to the nucleus, endoplasmic reticulum and tonoplast and was primarily expressed in the xylem and cambium. Overexpression of PdRabG3f in Populus alba × Populus glandulosa (84 K poplar) had inhibitory effects on vertical and radical growth. In the transgenic lines, there were evident changes in the levels of 15 gibberellin (GA) derivatives, and the application of exogenous GA3 partially restored the phenotypes mediated by GAs deficiency. The interaction between PdRabG3f and RIC4, which was the GA-responsive factor, provided additional explanation for PdRabG3f's inhibitory effect on poplar growth. RNA-seq analysis revealed differentially expressed genes (DEGs) associated with cell wall, xylem, and gibberellin response. PdRabG3f interfering endogenous GAs levels in poplar might involve the participation of MYBs and ultimately affected internode elongation and xylem development. This study provides a potential mechanism for gibberellin-mediated regulation of plant growth through Rab GTPases."
Authors: Yangang Pei, Qihan Xue, Peng Shu, Weijie Xu, Xiaofei Du, Mengbo Wu, Kaidong Liu, Julien Pirrello, Mondher Bouzayen, Yiguo Hong and Mingchun Liu.
Developmental Cell (2024)
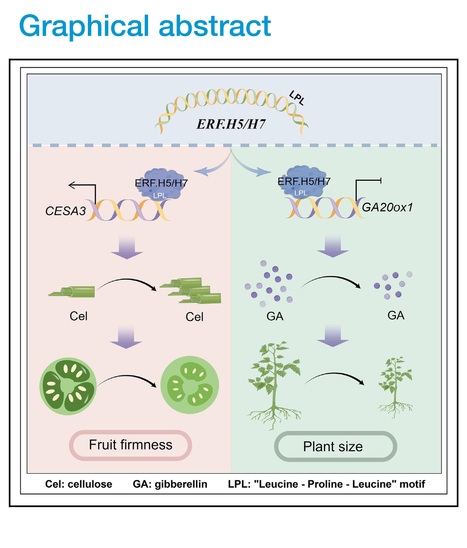
Editor's view: Pei et al. demonstrate the role of bifunctional transcription factors SlERF.H5 and SlERF.H7 in activating the cellulose biosynthesis gene SlCESA3 while repressing the gibberellin biosynthesis gene GA20ox1. The study highlights the distinct transcriptional regulatory functions of these ERFs in promoting cell wall formation and inhibiting plant growth.
Highlights • Cellulose is required for cell wall formation and firmness maintenance of fruits • SlERF.H5 and SlERF.H7 act as both transcriptional activators and repressors • The regulatory activity of SlERF.H5 and SlERF.H7 is mediated by a conserved motif
Abstract: "The plant cell wall is a dynamic structure that plays an essential role in development, but the mechanism regulating cell wall formation remains poorly understood. We demonstrate that two transcription factors, SlERF.H5 and SlERF.H7, control cell wall formation and tomato fruit firmness in an additive manner. Knockout of SlERF.H5, SlERF.H7, or both genes decreased cell wall thickness, firmness, and cellulose contents in fruits during early development, especially in double-knockout lines. Overexpressing either gene resulted in thicker cell walls and greater fruit firmness with elevated cellulose levels in fruits but severely dwarf plants with lower gibberellin contents. We further identified that SlERF.H5 and SlERF.H7 activate the cellulose biosynthesis gene SlCESA3 but repress the gibberellin biosynthesis gene GA20ox1. Moreover, we identified a conserved LPL motif in these ERFs responsible for their activities as transcriptional activators and repressors, providing insight into how bifunctional transcription factors modulate distinct developmental processes."
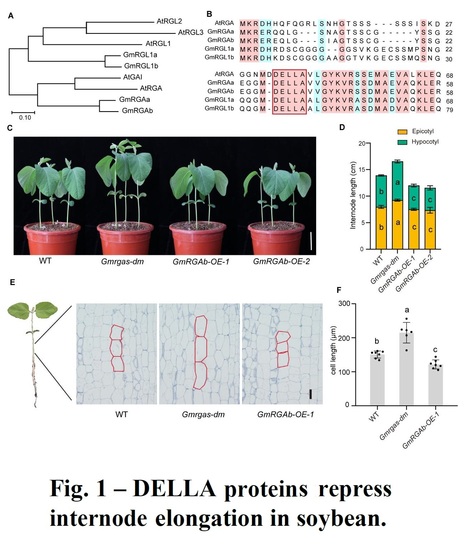
Authors: Zhuang Li, Qichao Tu, Xiangguang Lyu, Qican Cheng, Ronghuan Ji, Chao Qin, Jun Liu, Bin Liu, Hongyu Li and Tao Zhao.
The Crop Journal (2024)
Abstract: "Plant height influences plant architecture, lodging resistance, and yield performance. It is modulated by gibberellic acid (GA) metabolism and signaling. DELLA proteins, acting as central repressors of GA signaling, integrate various environmental and hormonal signals to regulate plant growth and development in Arabidopsis. We examined the role of two DELLA proteins, GmRGAa and GmRGAb, in soybean plant height control. Knockout of these proteins led to longer internodes and increased plant height, primarily by increasing cell elongation. GmRGAs functioned under different light conditions, including red, blue, and far-red light, to repress plant height. Interaction studies revealed that GmRGAs interacted with the blue light receptor GmCRY1b. Consistent with this, GmCRY1b partially regulated plant height via GmRGAs. Additionally, DELLA proteins were found to stabilize the protein GmSTF1/2, a key positive regulator of photomorphogenesis. This stabilization led to increased transcription of GmGA2ox-7b and subsequent reduction in plant height. This study enhances our understanding of DELLA-mediated plant height control, offering Gmrgaab mutants for soybean structure and yield optimization.
Authors: Jian You Wang, Guan-Ting Erica Chen, Aparna Balakrishna, Muhammad Jamil, Lamis Berqdar and Salim Al-Babili.
FEBS Letters (2024)
Summary: Previous study elucidated the alternative pathway of strigolactone (SL) biosynthesis, indicating the conversion of 9-cis-3-OH-β-apo-10′-carotenal into a 3-hydroxycarlactone in vitro. Herein, we fed wild-type (WT) and the SL-deficient d17 mutant rice seedlings with 13C-labeled 9-cis-3-OH-β-apo-10′-carotenal and analyzed their SLs by LC–MS/MS. Our results reveal that 9-cis-3-OH-β-apo-10′-carotenal is the SL precursor in planta, filling a knowledge gap in SL biosynthesis.
Abstract: "Strigolactones (SLs) play a crucial role in regulating plant architecture and mediating rhizosphere interactions. They are synthesized from all-trans-β-carotene converted into the intermediate carlactone (CL) via the intermediate 9-cis-β-apo-10′-carotenal. Recent studies indicate that plants can also synthesize 3-OH-CL from all-trans-β-zeaxanthin via the intermediate 9-cis-3-OH-β-apo-10′-carotenal. However, the question of whether plants can form bioactive SLs from 9-cis-3-OH-β-apo-10′-carotenal remains elusive. In this study, we supplied the 13C-labeled 9-cis-3-OH-β-apo-10′-carotenal to rice seedlings and monitored the synthesis of SLs using liquid chromatography-mass spectrometry (LC–MS) and Striga bioassay. We further validated the biological activity of 9-cis-3-OH-β-apo-10′-carotenal-derived SLs using the ccd7/d17 SL-deficient mutant, which demonstrated increased Striga seed-germinating activity and partial rescue of tiller numbers and plant height. Our results establish 9-cis-3-OH-β-apo-10′-carotenal as a significant SL biosynthetic intermediate with implications for understanding plant hormonal functions and potential applications in agriculture."
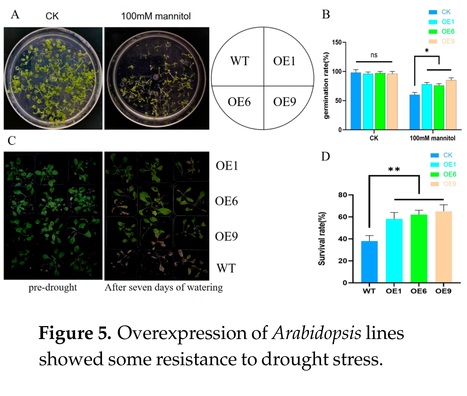
Authors: Chen Zhang, Fanhao Wang, Peng Jiao, Jiaqi Liu, Honglin Zhang, Siyan Liu, Shuyan Guan and Yiyong Ma.
International Journal of Molecular Sciences (2024)
Abstract: "Strigolactones (SLs) represent a recently identified class of plant hormones that are crucial for plant tillering and mycorrhizal symbiosis. The D14 gene, an essential receptor within the SLs signaling pathway, has been well-examined in crops, like rice (Oryza sativa L.) and Arabidopsis (Arabidopsis thaliana L.), yet the research on its influence in maize (Zea mays L.) remains scarce. This study successfully clones and establishes Arabidopsis D14 gene overexpression lines (OE lines). When compared with the wild type (WT), the OE lines exhibited significantly longer primary roots during germination. By seven weeks of age, these lines showed reductions in plant height and tillering, alongside slight decreases in rosette and leaf sizes, coupled with early aging symptoms. Fluorescence-based quantitative assays indicated notable hormonal fluctuations in OE lines versus the WT, implying that D14 overexpression disrupts plant hormonal homeostasis. The OE lines, exposed to cold, drought, and sodium chloride stressors during germination, displayed an especially pronounced resistance to drought. The drought resistance of OE lines, as evident from dehydration–rehydration assays, outmatched that of the WT lines. Additionally, under drought conditions, the OE lines accumulated less reactive oxygen species (ROS) as revealed by the assessment of the related physiological and biochemical parameters. Upon confronting the pathogens Pseudomonas syringae pv. tomato DC3000 (Pst DC3000), post-infection, fluorescence quantitative investigations showed a significant boost in the salicylic acid (SA)-related gene expression in OE lines compared to their WT counterparts. Overall, our findings designate the SL receptor D14 as a key upregulator of drought tolerance and a regulator in the biotic stress response, thereby advancing our understanding of the maize SL signaling pathway by elucidating the function of the pivotal D14 gene."
Authors: Jingye Cheng, Yong Jia, Camilla Hill, Tianhua He, Ke Wang, Ganggang Guo, Sergey Shabala, Meixue Zhou, Yong Han and Chengdao Li.
Journal of Advanced Research (2024)
Highlights • Genes concoding gibberellin 2-oxidase are expanded to ten copies in the barley genome and shown copy number variation in the barley pan genome. • The ten genes are transcriptionally expressed in a tissue-specific way, indicating divergent biological functions. • Functions of two genes of gibberellin 2-oxidase were validated by virus-induced gene silencing and CRISPR gene editing to control different agronomic traits. • A natural mutant for the gene was identified with semidwarf stature, early flowering and heavier seeds. • The results provide a promising strategy to minimise the adverse effects of the original Green Revolution semi-dwarf genes to develop the next generation of barley cultivars better adapted to a changing climate.
Abstract: "Introduction - Gibberellin (GA) is a vital phytohormone in regulating plant growth and development. During the “Green Revolution”, modification of GA-related genes created semi-dwarfing phenotype in cereal crops but adversely affected grain weight. Gibberellin 2-oxidases (GA2oxs) in barley act as key catabolic enzymes in deactivating GA, but their functions are still less known. Objectives - This study investigates the physiological function of two HvGA2ox genes in barley and identifies novel semi-dwarf alleles with minimum impacts on other agronomic traits. Methods - Virus-induced gene silencing and CRISPR/Cas9 technology were used to manipulate gene expression of HvGA2ox9 and HvGA2ox8a in barley and RNA-seq was conducted to compare the transcriptome between wild type and mutants. Also, field trials in multiple environments were performed to detect the functional haplotypes. Results - There were ten GA2oxs that distinctly expressed in shoot, tiller, inflorescence, grain, embryo and root. Knockdown of HvGA2ox9 did not affect plant height, while ga2ox8a mutants generated by CRISPR/Cas9 increased plant height and significantly altered seed width and weight due to the increased bioactive GA4 level. RNA-seq analysis revealed that genes involved in starch and sucrose metabolism were significantly decreased in the inflorescence of ga2ox8a mutants. Furthermore, haplotype analysis revealed one naturally occurring HvGA2ox8a haplotype was associated with decreased plant height, early flowering and wider and heavier seed. Conclusion - Our results demonstrate the potential of manipulating GA2ox genes to fine tune GA signalling and biofunctions in desired plant tissues and open a promising avenue for minimising the trade-off effects of Green Revolution semi-dwarfing genes on grain size and weight. The knowledge will promote the development of next generation barley cultivars with better adaptation to a changing climate."
Author: Ravi Gupta.
Plant Cell Reports (2024)
Abstract: "Key message - A recent study identified a natural deletion in the r-e-z haploblock which confers a semi-dwarf trait, higher nitrogen use efficiency, and improved yield in semi-dwarf wheat varieties by attenuating the brassinosteroid signaling."
Authors: Pierre Gautrat, Sara Buti, Andrés Romanowski, Guido Buijs and Ronald Pierik.
bioRxiv (2023)
Abstract: "Plants growing at high densities can detect competitors through changes in the composition of light reflected by neighbours. In response to this far-red-enriched light, plants elicit adaptive shade avoidance responses for light capture, but these need to be balanced against other input signals, such as nutrient availability. Here, we used a combination of transcriptomics, growth assays and dedicated genetic and pharmacological interventions to demonstrate how Arabidopsis integrates light and nitrate signalling. We unveiled that nitrate modulates shade avoidance via a previously unknown shade response pathway that involves root-derived transZeatin (tZ). Under nitrate-sufficient conditions, tZ promotes hypocotyl elongation specifically in the presence of supplemental far-red light. This occurs via PIF transcription factors-dependent inhibition of type-A ARRs cytokinin response inhibitors. Our data thus reveal how plants co-regulate responses to shade cues with root-derived information about nutrient availability, and how they restrict responses to this information to specific light conditions in the shoot."
Author: Carlisle Bascom, Jr.
The Plant Cell (2024)
Excerpts: "Myriad genes control plant size and development in Arabidopsis (A. thaliana), including those encoding the BASIC PENTACYSTEINE (BPC) class of transcription factors (Monfared et al. 2011). However, whether BPCs regulate development in woody plants is an open question. In this issue, Haiyan Zhao and colleagues (Zhao et al. 2024) reveal some details of the genetic regulation of dwarfism in apple trees, Malus domestica. Through genetics, bioinformatics, and biochemical assays, Zhao and colleagues elucidated a key molecular mechanism for plant dwarfism (see Figure)."
"Histone trimethylation (H3K27me3) results in repression of target genes (Cai et al. 2021). Here, the authors found that H3K27me3 enrichment was significantly increased at MdYUC2a and 6b loci in OX plants. H3K27me3 modifications are facilitated by a large complex of proteins known as the polycomb group. The author used biochemical techniques to demonstrate that MdBPC2 interacts with the polycomb group member LIKE HETEROCHROMATIN PROTEIN1 (LHP1). Indeed, MdYUC2a and 6b loci are enriched with MdLHP1 (see Figure). With this result, the authors proposed a straight-forward model whereby MdBPC2 protein recruits LHP1 protein to the promoters of a subset of YUCCA genes to repress their expression, thereby reducing the amount of auxin produced. In dwarf rootstocks, increased MdBPC2 expression results in much less MdYUC2a and 6b enzymes, and shorter, auxin-deficient plants (see Figure). Moving forward, one can see BPCs as an attractive target for crop breeding programs where dwarf plants are the goal."
Authors: Canran Sun, Yang Liu, Guofang Li, Yanle Chen, Mengyuan Li, Ruihua Yang, Yongtian Qin, Yongqiang Chen, Jinpeng Cheng, Jihua Tang and Zhiyuan Fu.
The Crop Journal (2024)
Abstract: "Plant height (PH) is associated with lodging resistance and planting density, which is regulated by a complicated gene network. In this study, we identified a spontaneous dwarfing mutation in maize, m30, with decreased internode number and length but increased internode diameter. A candidate gene, ZmCYP90D1, which encodes a member of the cytochrome P450 family, was isolated by map-based cloning. ZmCYP90D1 was constitutively expressed and showed highest expression in basal internodes, and its protein was targeted to the nucleus. A G-to-A substitution was identified to be the causal mutation, which resulted in a truncated protein in m30. Loss of function of ZmCYP90D1 changed expression of hormone-responsive genes, in particular brassinosteroid (BR)-responsive genes which is mainly involved in cell cycle regulation and cell wall extension and modification in plants. The concentration of typhasterol (TY), a downstream intermediate of ZmCYP90D1 in the BR pathway, was reduced. A haplotype conferring dwarfing without reducing yield was identified. ZmCYP90D1 was inferred to influence plant height and stalk diameter via hormone-mediated cell division and cell growth via the BR pathway."
|


 Your new post is loading...
Your new post is loading...