Your new post is loading...
 Your new post is loading...
Authors: Gang Wang, Xi Chen, Chengzhi Yu, Xiaobao Shi, Wenxian Lan, Chaofeng Gao, Jun Yang, Huiling Dai, Xiaowei Zhang, Huili Zhang, Boyu Zhao, Qi Xie, Nan Yu, Zuhua He, Yu Zhang and Ertao Wang.
Nature (2024)
One-sentence summary: The ubiquitin E3 ligase OsCIE1 acts as a brake to inhibit OsCERK1 during homeostasis; this brake is released after chitin stimulation.
Abstract: "Plant pattern-recognition receptors perceive microorganism-associated molecular patterns to activate immune signalling1,2. Activation of the pattern-recognition receptor kinase CERK1 is essential for immunity, but tight inhibition of receptor kinases in the absence of pathogen is crucial to prevent autoimmunity3,4. Here we find that the U-box ubiquitin E3 ligase OsCIE1 acts as a molecular brake to inhibit OsCERK1 in rice. During homeostasis, OsCIE1 ubiquitinates OsCERK1, reducing its kinase activity. In the presence of the microorganism-associated molecular pattern chitin, active OsCERK1 phosphorylates OsCIE1 and blocks its E3 ligase activity, thus releasing the brake and promoting immunity. Phosphorylation of a serine within the U-box of OsCIE1 prevents its interaction with E2 ubiquitin-conjugating enzymes and serves as a phosphorylation switch. This phosphorylation site is conserved in E3 ligases from plants to animals. Our work identifies a ligand-released brake that enables dynamic immune regulation."
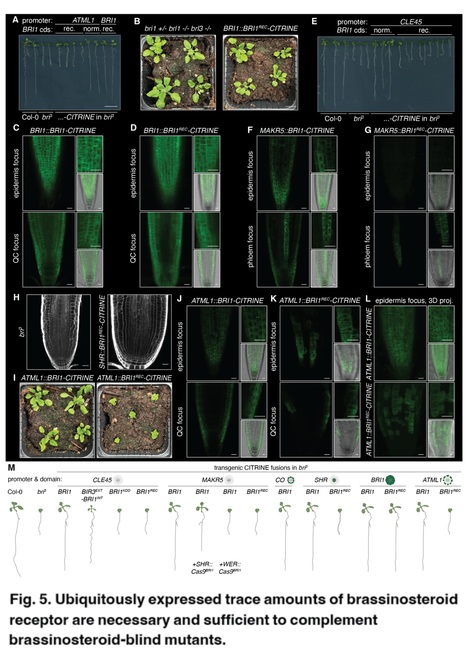
Authors: Noel Blanco-Touriñán, Surbhi Rana, Trevor M. Nolan, Kunkun Li, Nemanja Vukašinović, Che-Wei Hsu, Eugenia Russinova and Christian S. Hardtke.
bioRxiv (2024)
Abstract: "Brassinosteroid signaling is essential for plant growth as exemplified by the dwarf phenotype of loss-of-function mutants in BRASSINOSTEROID INSENSITIVE 1 (BRI1), a ubiquitously expressed Arabidopsis brassinosteroid receptor gene. Complementation of brassinosteroid-blind receptor mutants by BRI1 expression with various tissue-specific promoters implied that local brassinosteroid signaling may instruct growth non-cell-autonomously. Here we performed such rescues with a panel of receptor variants and promoters, in combination with tissue-specific transgene knockouts. Our experiments demonstrate that brassinosteroid receptor expression in several tissues is necessary but not sufficient for rescue. Moreover, complementation with tissue-specific promoters requires the genuine BRI1 gene body sequence, which confers ubiquitous expression of trace receptor amounts that are sufficient to promote brassinosteroid-dependent root growth. Our data, therefore, argue for a largely cell-autonomous action of brassinosteroid receptors."
Authors: J. S. Chen, S. T. Wang, Q. Mei, T. Sun, J. T. Hu, G. S. Xiao, H. Chen and Y. H. Xuan.
Plant Molecular Biology (2024)
Abstract: "Plants have a variety of regulatory mechanisms to perceive, transduce, and respond to biotic and abiotic stress. One such mechanism is the calcium-sensing CBL–CIPK system responsible for the sensing of specific stressors, such as drought or pathogens. CBLs perceive and bind Calcium (Ca2+) in response to stress and then interact with CIPKs to form an activated complex. This leads to the phosphorylation of downstream targets, including transporters and ion channels, and modulates transcription factor levels and the consequent levels of stress-associated genes. This review describes the mechanisms underlying the response of the CBL–CIPK pathway to biotic and abiotic stresses, including regulating ion transport channels, coordinating plant hormone signal transduction, and pathways related to ROS signaling. Investigation of the function of the CBL–CIPK pathway is important for understanding plant stress tolerance and provides a promising avenue for molecular breeding."
Authors: Feimei Guo, Minghui Lv, Jingjie Zhang and Jia Li.
Plant and Cell Physiology (2024)
Abstract: "Brassinosteroids (BRs) are a group of polyhydroxylated phytosterols that play essential roles in regulating plant growth and development as well as stress adaptation. It is worth noting that BRs do not function alone, but rather they crosstalk with other endogenous signaling molecules, including the phytohormones auxin, cytokinins (CKs), gibberellins (GAs), abscisic acid (ABA), ethylene (ET), jasmonates (JAs), salicylic acid (SA), and strigolactones (SLs), forming elaborate signaling networks to modulate plant growth and development. BRs interact with other phytohormones mainly by regulating each others’ homeostasis, transport, or signaling pathway at the transcriptional and posttranslational levels. In this review, we focus our attention on current research progress in BR signal transduction and the crosstalk between BRs and other phytohormones."
Authors: Jorge Hernández-García, Antonio Serrano-Mislata, María Lozano-Quiles, Cristina Úrbez, María A. Nohales, Noel Blanco-Touriñán, Huadong Peng, Rodrigo Ledesma-Amaro and Miguel A. Blázquez.
PNAS (2024)
Significance: DELLA proteins are plant-specific transcriptional hubs integrating environmental signals with endogenous cues. In order to regulate downstream processes, DELLAs modulate the activity of hundreds of transcription factors (TFs) and transcriptional regulators in various ways. Here, we describe the molecular mechanism underlying DELLA coactivator function. We show that DELLAs act as transcriptional activators by interacting with the Mediator complex subunit MED15. This interaction is necessary to regulate a specific subset of DELLA-dependent responses that are mediated by transcriptional coactivation, but not those regulated by TF sequestration. We further show that this mechanism is present in bryophyte DELLAs and thus represents a conserved mechanism of DELLA function in land plants.
Abstract: "DELLA proteins are negative regulators of the gibberellin response pathway in angiosperms, acting as central hubs that interact with hundreds of transcription factors (TFs) and regulators to modulate their activities. While the mechanism of TF sequestration by DELLAs to prevent DNA binding to downstream targets has been extensively documented, the mechanism that allows them to act as coactivators remains to be understood. Here, we demonstrate that DELLAs directly recruit the Mediator complex to specific loci in Arabidopsis, facilitating transcription. This recruitment involves DELLA amino-terminal domain and the conserved MED15 KIX domain. Accordingly, partial loss of MED15 function mainly disrupted processes known to rely on DELLA coactivation capacity, including cytokinin-dependent regulation of meristem function and skotomorphogenic response, gibberellin metabolism feedback, and flavonol production. We have also found that the single DELLA protein in the liverwort Marchantia polymorpha is capable of recruiting MpMED15 subunits, contributing to transcriptional coactivation. The conservation of Mediator-dependent transcriptional coactivation by DELLA between Arabidopsis and Marchantia implies that this mechanism is intrinsic to the emergence of DELLA in the last common ancestor of land plants."
Authors: Amanpreet Kaur, Norman B. Best, Thomas Hartwig, Josh Budka, Rajdeep Khangura, Steven McKenzie, Alejandro Aragón-Raygoza, Josh Strable, Burkhard Schulz and Brian P. Dilkes.
Plant Physiology (2023)
One-sentence summary: Molecular identity of a maize semidwarf mutant reveals a role for the maize GRAS domain transcription factor ortholog of DWARF AND LOW TILLERING in brassinosteroid signaling.
Abstract: "Brassinosteroids (BR) and gibberellins (GA) regulate plant height and leaf angle in maize (Zea mays). Mutants with defects in BR or GA biosynthesis or signaling identify components of these pathways and enhance our knowledge about plant growth and development. In this study, we characterized three recessive mutant alleles of GRAS transcription factor 42 (gras42) in maize, a GRAS transcription factor gene orthologous to the DWARF AND LOW TILLERING (DLT) gene of rice (Oryza sativa). These maize mutants exhibited semi-dwarf stature, shorter and wider leaves, and more upright leaf angle. Transcriptome analysis revealed a role for GRAS42 as a determinant of BR signaling. Analysis of the expression consequences from loss of GRAS42 in the gras42-mu1021149 mutant indicated a weak loss of BR signaling in the mutant, consistent with its previously demonstrated role in BR signaling in rice. Loss of BR signaling was also evident by the enhancement of weak BR biosynthetic mutant alleles in double mutants of nana plant1-1 and gras42-mu1021149. The gras42-mu1021149 mutant had little effect on GA-regulated gene expression, suggesting that GRAS42 is not a regulator of core GA signaling genes in maize. Single cell expression data identified gras42 expressed among cells in the G2/M phase of the cell cycle consistent with its previously demonstrated role in cell cycle gene expression in Arabidopsis (Arabidopsis thaliana). Cis-acting natural variation controlling GRAS42 transcript accumulation was identified by expression genome-wide association study (eGWAS) in maize. Our results demonstrate a conserved role for GRAS42/SCARECROW-LIKE 28 (SCL28)/DLT in BR signaling, clarify the role of this gene in GA signaling, and suggest mechanisms of tillering and leaf angle control by BR."
Authors: Lei Liang and Xiangyang Hu.
Phyton (2024)
Abstract: "With the rapid development of modern molecular biology and bioinformatics, many studies have proved that transcription factors play an important role in regulating the growth and development of plants. SPATULA (SPT) belongs to the bHLH transcription family and participates in many processes of regulating plant growth and development. This review systemically summarizes the multiple roles of SPT in plant growth, development, and stress response, including seed germination, flowering, leaf size, carpel development, and root elongation, which is helpful for us to better understand the functions of SPT."
Authors: André Kessler and Michael B. Mueller.
Plant Signaling & Behavior (2024)
Abstract: "Plant induced responses to environmental stressors are increasingly studied in a behavioral ecology context. This is particularly true for plant induced responses to herbivory that mediate direct and indirect defenses, and tolerance. These seemingly adaptive alterations of plant defense phenotypes in the context of other environmental conditions have led to the discussion of such responses as intelligent behavior. Here we consider the concept of plant intelligence and some of its predictions for chemical information transfer in plant interaction with other organisms. Within this framework, the flow, perception, integration, and storage of environmental information are considered tunable dials that allow plants to respond adaptively to attacking herbivores while integrating past experiences and environmental cues that are predictive of future conditions. The predictive value of environmental information and the costs of acting on false information are important drivers of the evolution of plant responses to herbivory. We identify integrative priming of defense responses as a mechanism that allows plants to mitigate potential costs associated with acting on false information. The priming mechanisms provide short- and long-term memory that facilitates the integration of environmental cues without imposing significant costs. Finally, we discuss the ecological and evolutionary prediction of the plant intelligence hypothesis."
Authors: Xuehuan Feng, Jinfang Zheng, Iker Irisarri, Huihui Yu, Bo Zheng, Zahin Ali, Sophie de Vries, Jean Keller, Janine M. R. Fürst-Jansen, Armin Dadras, Jaccoline M. S. Zegers, Tim P. Rieseberg, Amra Dhabalia Ashok, Tatyana Darienko, Maaike J. Bierenbroodspot, Lydia Gramzow, Romy Petroll, Fabian B. Haas, Noe Fernandez-Pozo, Orestis Nousias, Tang Li, Elisabeth Fitzek, W. Scott Grayburn, Nina Rittmeier, Charlotte Permann, Florian Rümpler, John M. Archibald, Günter Theißen, Jeffrey P. Mower, Maike Lorenz, Henrik Buschmann, Klaus von Schwartzenberg, Lori Boston, Richard D. Hayes, Chris Daum, Kerrie Barry, Igor V. Grigoriev, Xiyin Wang, Fay-Wei Li, Stefan A. Rensing, Julius Ben Ari, Noa Keren, Assaf Mosquna, Andreas Holzinger, Pierre-Marc Delaux, Chi Zhang, Jinling Huang, Marek Mutwil, Jan de Vries and Yanbin Yin
Nature Genetics (2024)
Editor's view: Genome assemblies of four filamentous Zygnematophyceae and co-expression network analyses shed light on the evolutionary roots of the mechanism for balancing environmental responses and multicellular growth.
Abstract: "Zygnematophyceae are the algal sisters of land plants. Here we sequenced four genomes of filamentous Zygnematophyceae, including chromosome-scale assemblies for three strains of Zygnema circumcarinatum. We inferred traits in the ancestor of Zygnematophyceae and land plants that might have ushered in the conquest of land by plants: expanded genes for signaling cascades, environmental response, and multicellular growth. Zygnematophyceae and land plants share all the major enzymes for cell wall synthesis and remodifications, and gene gains shaped this toolkit. Co-expression network analyses uncover gene cohorts that unite environmental signaling with multicellular developmental programs. Our data shed light on a molecular chassis that balances environmental response and growth modulation across more than 600 million years of streptophyte evolution."
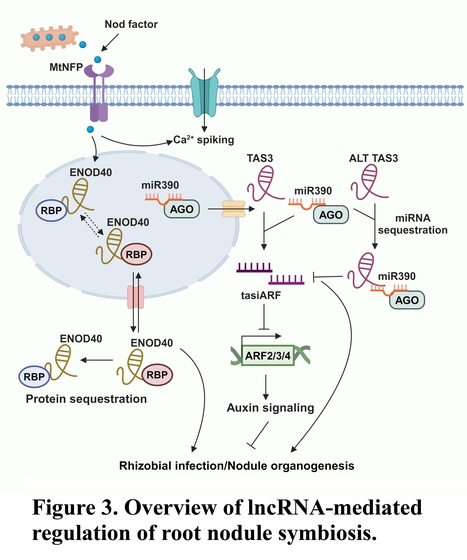
Authors: Muhammad Fahad, Leeza Tariq, Sajid Muhammad and Liang Wu.
Plant Communications (2024)
Abstract: "Long non-coding RNAs (lncRNAs) have emerged as integral gene expression regulators underlying plant growth, development, and adaptation. To adapt to the heterogeneous and dynamic rhizosphere, plants use interconnected regulatory mechanisms to optimally fine-tune gene expression governing interactions with soil biota, nutrient acquisition, and heavy metal tolerance. Recently, high-throughput sequencing has enabled the identification of plant lncRNAs responsive to rhizosphere biotic and abiotic cues. Here, we examine lncRNA biogenesis, classification, and mode of action, highlighting the functions of lncRNAs in mediating plant adaptation to diverse rhizosphere factors. We then discuss studies that revealed lncRNA significance and target genes during developmental plasticity and stress responses at the rhizobium interface. Thus, a comprehensive understanding of specific lncRNAs, their regulatory targets, and the intricacies of their functional interaction networks will provide crucial insights into how these transcriptomic switches fine-tune responses to shifting rhizosphere signals. As we look ahead, we foresee that single-cell dissection of cell type-specific lncRNA regulatory dynamics will enhance our understanding of precise developmental modulation mechanisms enabling plant rhizosphere adaptation. Overcoming future challenges through multi-omics and genetic approaches will better reveal the integral lncRNA roles governing plant adaptation to the belowground environment."
Authors: Ping Yun, Cengiz Kaya and Sergey Shabala.
The Crop Journal (2024)
Abstract: "Salinity stress is a major environmental stress affecting crop productivity, and its negative impact on global food security is only going to increase, due to current climate trends. Salinity tolerance was present in wild crop relatives but significantly weakened during domestication. Regaining it back requires a good understanding of molecular mechanisms and traits involved in control of plant ionic and ROS homeostasis. This review summarizes our current knowledge on the role of major plant hormones (auxin, cytokinins, abscisic acid, salicylic acid, and jasmonate) in plants adaptation to soil salinity. We firstly discuss the role of hormones in controlling root tropisms, root growth and architecture (primary root elongation, meristematic activity, lateral root development, and root hairs formation). Hormone-mediated control of uptake and sequestration of key inorganic ions (sodium, potassium, and calcium) is then discussed followed by regulation of cell redox balance and ROS signaling in salt-stressed roots. Finally, the role of epigenetic alterations such as DNA methylation and histone modifications in control of plant ion and ROS homeostasis and signaling is discussed. This data may help develop novel strategies for breeding and cultivating salt-tolerant crops and improving agricultural productivity in saline regions."
Authors: Isabel Cristina Vélez-Bermúdez and Wolfgang Schmidt.
Nature Plants (2024)
Summary: "The functions of a small family of non-secreted peptides, originally identified as critical communicators of the plant’s iron status, have expanded. The involvement of these effectors in disparate signalling cascades underlines the pivotal role peptides have in responses to the environment."
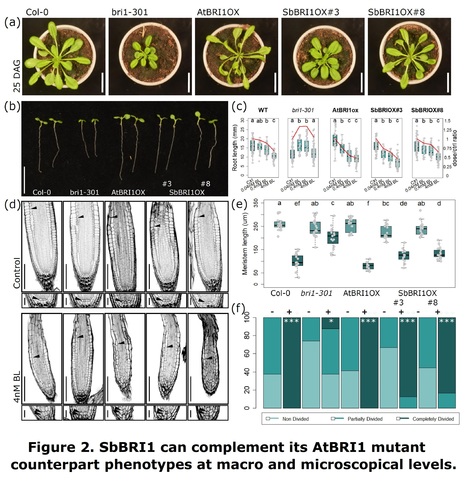
Authors: Andrés Rico-Medina, David Blasco-Escámez, Juan B. Fontanet-Manzaneque, Natalie Laibach, Fidel Lozano-Elena, Damiano Martignago and Ana I. Caño-Delgado.
bioRxiv (2024)
Abstract: "- The high sequence and structural similarities between BRI1 brassinosteroid receptors of Arabidopsis (AtBRI1) and sorghum (SbBRI1) prompted us to study the functionally conserved roles of BRI in both organisms. - Introducing sorghum SbBRI1 in Arabidopsis bri1 mutants restores defective growth and developmental phenotypes to WT levels. - Sorghum mutants for SbBRI1 receptors show defective BR sensitivity and impaired growth and development throughout the entire sorghum life cycle. Embryonic analysis of sorghum primary roots permit to trace back root growth and development to early stages, revealing the functionally conserved roles of SbBRI1 receptor in BR perception during meristem development. RNA-seq analysis uncovers the downstream regulation of the SbBRI1 pathway in cell wall biogenesis during cell growth. - Together, these results uncover that sorghum SbBRI1 receptor protein play functionally conserved roles in plant growth and development, while encourage the study of BR pathways in sorghum and its implications for improving resilience in cereal crops."
|
Authors: Lisa Bohn, Jin Huang, Susan Weidig, Zhenyu Yang, Christoph Heidersberger, Bernard Genty, Pascal Falter-Braun, Alexander Christmann and Erwin Grill.
Nature (2024)
One-sentence summary: TWA1 is a temperature-sensing transcriptional co-regulator that is needed for basal and acquired thermotolerance in Arabidopsis thaliana.
Abstract: "Plants exposed to incidences of excessive temperatures activate heat-stress responses to cope with the physiological challenge and stimulate long-term acclimation1,2. The mechanism that senses cellular temperature for inducing thermotolerance is still unclear3. Here we show that TWA1 is a temperature-sensing transcriptional co-regulator that is needed for basal and acquired thermotolerance in Arabidopsis thaliana. At elevated temperatures, TWA1 changes its conformation and allows physical interaction with JASMONATE-ASSOCIATED MYC-LIKE (JAM) transcription factors and TOPLESS (TPL) and TOPLESS-RELATED (TPR) proteins for repressor complex assembly. TWA1 is a predicted intrinsically disordered protein that has a key thermosensory role functioning through an amino-terminal highly variable region. At elevated temperatures, TWA1 accumulates in nuclear subdomains, and physical interactions with JAM2 and TPL appear to be restricted to these nuclear subdomains. The transcriptional upregulation of the heat shock transcription factor A2 (HSFA2) and heat shock proteins depended on TWA1, and TWA1 orthologues provided different temperature thresholds, consistent with the sensor function in early signalling of heat stress. The identification of the plant thermosensors offers a molecular tool for adjusting thermal acclimation responses of crops by breeding and biotechnology, and a sensitive temperature switch for thermogenetics."
Authors: Byron Rusnak, Frances K. Clark, Batthula Vijaya Lakshmi Vadde and Adrienne H.K. Roeder.
Annual Review of Cell and Developmental Biology (2024)
Abstract: "One of the fundamental questions in developmental biology is how a cell is specified to differentiate as a specialized cell type. Traditionally, plant cell types were defined based on their function, location, morphology, and lineage. Currently, in the age of single-cell biology, researchers typically attempt to assign plant cells to cell types by clustering them based on their transcriptomes. However, because cells are dynamic entities that progress through the cell cycle and respond to signals, the transcriptome also reflects the state of the cell at a particular moment in time, raising questions about how to define a cell type. We suggest that these complexities and dynamics of cell states are of interest and further consider the roles signaling, stochasticity, cell cycle, and mechanical forces play in plant cell fate specification. Once established, cell identity must also be maintained. With the wealth of single-cell data coming out, the field is poised to elucidate both the complexity and dynamics of cell states."
Author: Peter V. Minorsky.
Plant Signaling & Behavior (2024)
Abstract: "The 21st-century “plant neurobiology” movement is an amalgam of scholars interested in how “neural processes”, broadly defined, lead to changes in plant behavior. Integral to the movement (now called plant behavioral biology) is a triad of historically marginalized subdisciplines, namely plant ethology, whole plant electrophysiology and plant comparative psychology, that set plant neurobiology apart from the mainstream. A central tenet held by these “triad disciplines” is that plants are exquisitely sensitive to environmental perturbations and that destructive experimental manipulations rapidly and profoundly affect plant function. Since destructive measurements have been the norm in plant physiology, much of our “textbook knowledge” concerning plant physiology is unrelated to normal plant function. As such, scientists in the triad disciplines favor a more natural and holistic approach toward understanding plant function. By examining the history, philosophy, sociology and psychology of the triad disciplines, this paper refutes in eight ways the criticism that plant neurobiology presents nothing new, and that the topics of plant neurobiology fall squarely under the purview of mainstream plant physiology. It is argued that although the triad disciplines and mainstream plant physiology share the common goal of understanding plant function, they are distinct in having their own intellectual histories and epistemologies."
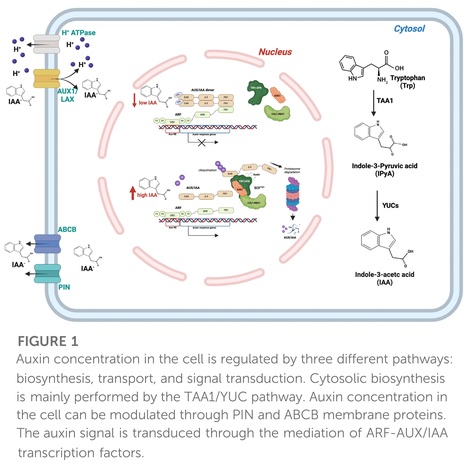
Authors: Davide Marzi, Patrizia Brunetti, Shashank Sagar Saini, Gitanjali Yadav, Giuseppe Diego Puglia and Raffaele Dello Ioio.
Frontiers in Genetics (2024)
Abstract: "Global climate change (GCC) is posing a serious threat to organisms, particularly plants, which are sessile. Drought, salinity, and the accumulation of heavy metals alter soil composition and have detrimental effects on crops and wild plants. The hormone auxin plays a pivotal role in the response to stress conditions through the fine regulation of plant growth. Hence, rapid, tight, and coordinated regulation of its concentration is achieved by auxin modulation at multiple levels. Beyond the structural enzymes involved in auxin biosynthesis, transport, and signal transduction, transcription factors (TFs) can finely and rapidly drive auxin response in specific tissues. Auxin Response Factors (ARFs) such as the ARF4, 7, 8, 19 and many other TF families, such as WRKY and MADS, have been identified to play a role in modulating various auxin-mediated responses in recent times. Here, we review the most relevant and recent literature on TFs associated with the regulation of the biosynthetic, transport, and signalling auxin pathways and miRNA-related feedback loops in response to major abiotic stresses. Knowledge of the specific role of TFs may be of utmost importance in counteracting the effects of GCC on future agriculture and may pave the way for increased plant resilience."
Authors: Bihai Shi, Amelia Felipo-Benavent, Guillaume Cerutti, Carlos Galvan-Ampudia, Lucas Jilli, Geraldine Brunoud, Jérome Mutterer, Elody Vallet, Lali Sakvarelidze-Achard, Jean-Michel Davière, Alejandro Navarro-Galiano, Ankit Walia, Shani Lazary, Jonathan Legrand, Roy Weinstain, Alexander M. Jones, Salomé Prat, Patrick Achard and Teva Vernoux.
Nature Communications (2024)
Editor's view: Engineering of a biosensor allows the authors to map the signaling activity of the phytohormones gibberellins (GAs) and to show that GAs orient cell division at the shoot apex to establish the organization in parallel cell files of plant stems.
Abstract: "Growth at the shoot apical meristem (SAM) is essential for shoot architecture construction. The phytohormones gibberellins (GA) play a pivotal role in coordinating plant growth, but their role in the SAM remains mostly unknown. Here, we developed a ratiometric GA signaling biosensor by engineering one of the DELLA proteins, to suppress its master regulatory function in GA transcriptional responses while preserving its degradation upon GA sensing. We demonstrate that this degradation-based biosensor accurately reports on cellular changes in GA levels and perception during development. We used this biosensor to map GA signaling activity in the SAM. We show that high GA signaling is found primarily in cells located between organ primordia that are the precursors of internodes. By gain- and loss-of-function approaches, we further demonstrate that GAs regulate cell division plane orientation to establish the typical cellular organization of internodes, thus contributing to internode specification in the SAM."
Authors: Tomás Urzúa Lehuedé, Victoria Berdion Gabarain, Tomas Moyano, Lucia Ferrero, Miguel Angel Ibeas, Hernan Salinas-Grenet, Gerardo Núñez-Lillo, Romina Acha, Jorge Perez, Florencia Perotti, Virginia Natali Miguel, Fiorella Paola Spies, Miguel A. Rosas, Michitaro Shibata, Diana R. Rodríguez-García, Adrian A. Moreno, Keiko Sugimoto, Karen Sanguinet, Claudio Meneses, Raquel L. Chan, Federico Ariel, Jose M. Alvarez and José M. Estevez.
bioRxiv (2024)
Abstract: "The root hair (RH) cells can elongate to several hundred times their initial size, and are an ideal model system for investigating cell size control. Their development is influenced by both endogenous and external signals, which are combined to form a integrative response. Surprisingly, a low temperature condition of 10°C causes an increased RH growth in Arabidopsis and in several monocots, even when the development of the rest of the root and aerial parts of the plant are halted. Previously, we demonstrated a strong correlation between the growth response and a significant decrease in nutrient availability in the medium under low temperature conditions. However, the molecular basis responsible for receiving and transmitting signals related to the availability of nutrients in the soil, and their relation to plant development, remain largely unknown. We decided to further investigate the intricate molecular processes behind the particular responsiveness of this root cell type at low temperature. In this study, we have discovered a gene regulatory network (GRN) controlling early transcriptome responses to low temperature. This GNR is commanded by specific transcription factors (FTs), namely ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), a member of the homeodomain leucine zipper (HD-Zip I) group I 13 (AtHB13), the trihelix TF GT2-LIKE1 (GTL1), and a previously unidentified MYB-like TF (AT2G01060). Furthermore, we have identified four downstream RSL4 targets AtHB16, AtHB23, EARLY-RESPONSIVE TO DEHYDRATION 7 (ERD7) and ERD10 suggesting their participation in the regulation of RH development under these conditions. Functional analysis shows that such components of the RSL4-dependent transcriptional cascade influence the subsequent RH growth response to low temperature. These discoveries enhance our comprehension of how plants synchronize the RH growth in response to variations in temperature and nutrient availability at the cellular level."
Authors: Rina Saito, Yuho Nishizato, Tsumugi Kitajima, Misuzu Nakayama, Yousuke Takaoka, Nobuki Kato and Minoru Ueda.
bioRxiv (2024)
Abstract: "(+)-cis-12-oxo-phytodienoic acid (cis-OPDA) is a biosynthetic precursor of the plant hormone (+)-7-iso-jasmonoyl-L-isoleucine (JA-Ile). It functions as an endogenous chemical signal independent of the JA-Ile receptor COI1-JAZ in Arabidopsis thaliana. The bioactive form of cis-OPDA that induces COI1-JAZ-independent gene expression remains unknown. In this study, we hypothesized that the genuine bioactive forms of cis-OPDA are the downstream metabolites, which upregulate the expression of the OPDA marker genes such as ZAT10/ERF5 in a JA-Ile-independent manner. These downstream metabolites function independently of the JA-Ile-COI1-JAZ-MYCs canonical jasmonate signaling module, and its electrophilic nature is essential for its bioactivity."
Authors: Yuming Sun and Alisdair R. Fernie.
Trends in Plant Science (2024)
Highlights Plant secondary metabolism plays a paramount role in plant adaptation to global climate change, a comprehensive understanding of the interplay between plant secondary metabolism and climate variations favors crop improvement in a climate-fluctuating world. Climate changes exert diverse effects on plant secondary metabolism, determined by factors such as metabolite type, stress intensity, and plant species. Combinations of two individual climatic factors can result in an amplifying, neutralizing, or one-factor-dominated consequence, but our knowledge of these consequences is limited. Resource allocation and growth-defense trade-off hypothesis partially explain how plants respond to climate changes through secondary metabolism at the ecological level, while hormone signaling, hormones, and transcriptional and post-transcriptional regulation networks are involved with the molecular mechanisms underlying these responses.
Abstract: "Climate changes have unpredictable effects on ecosystems and agriculture. Plants adapt metabolically to overcome these challenges, with plant secondary metabolites (PSMs) being crucial for plant–environment interactions. Thus, understanding how PSMs respond to climate change is vital for future cultivation and breeding strategies. Here, we review PSM responses to climate changes such as elevated carbon dioxide, ozone, nitrogen deposition, heat and drought, as well as a combinations of different factors. These responses are complex, depending on stress dosage and duration, and metabolite classes. We finally identify mechanisms by which climate change affects PSM production ecologically and molecularly. While these observations provide insights into PSM responses to climate changes and the underlying regulatory mechanisms, considerable further research is required for a comprehensive understanding."
Authors: Pragya Yadav, Shashank Kumar Yadav, Meenu Singh, Madan Pal Singh and Viswanathan Chinnusamy.
Plant Physiology Reports (2024)
Abstract: "Rice (Oryza sativa L.) is one of the key food crops in ensuring global food security and accounts for nearly one fourth of the world’s total food supply. Rice production is significantly affected by abiotic stresses. Cytokinins (CKs) play a pivotal role in the regulation of the plant growth, development and abiotic stress responses. In present study, genome-wise analysis led to the identification of 10 Isopentenyl transferase (IPT) gene family members, encoding rate-limiting enzymes in CK biosynthesis pathway in rice. Phylogenetic analysis categorized IPTs into four subfamilies that are structurally and functionally conserved across species with similar gene structures, motifs, domains, cis-acting elements were identified. We performed a genome-wide identification of IPT family members from the rice genome, identified their chromosomal location, gene structure, protein properties, cis-element and phylogeny. Further, Real-time PCR analysis was carried out to understand the spatio-temporal and stress responsive expression of IPT gene family members. OsIPT1, OsIPT2, OsIPT4, OsIPT7, OsIPT8 and OsIPT10 showed similar expression level in both root and shoot of seedling or more expression in root, while OsIPT3, OsIPT5, OsIPT6 and OsIPT9 showed higher expression in shoot as compared with root in seedlings. Abiotic stress responsive expression analysis showed that abiotic stresses reduced the expression of all family members except IPT6. Analysis of expression of IPTs in rice cv. Kitaake (low biomass and low yielding) and Taipei (high biomass and high yielding) showed that expression of IPTs were higher in different tissues in Taipei. Present study has also elaborately described the tissue and stage specific expression and for further analysis of IPT in the regulation of biotic and abiotic stress resistance, growth, and development in rice, and provides a comprehensive analysis of the OsIPT gene family, including structures and functions."
Authors: Jiangzhe Zhao, Jingqi Wang, Jie Liu, Penghong Zhang, Guzel Kudoyarova, Chang-Jun Liu and Kewei Zhang.
Plant Communications (2024)
Abstract: "Cytokinins are a type of mobile phytohormone that regulate plant growth, development, and environmental adaptability. The major cytokinin species include isopentenyl adenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DZ). The spatial distributions of different cytokinin species in different organelles, cells, tissues, and organs are primarily shaped by biosynthesis via isopentenyltransferases (IPT), cytochrome P450 monooxygenase, and 5’-ribonucleotide phosphohydrolase, and by conjugation or catabolism via glycosyltransferase or cytokinin oxidase/dehydrogenase (CKX). Cytokinins bind to histidine receptor kinases (HKs) in the endoplasmic reticulum (ER) or plasma membrane (PM) and relay signals to response regulators (RRs) in the nucleus by shuttle proteins known as histidine phosphotransfer proteins (HPs). The movements of cytokinins from sites of biosynthesis to signal perception sites usually require long-distance, intercellular, and intracellular transport. In the past decade, ATP-binding cassette (ABC) transporters, purine permeases (PUP), AZA-GUANINE RESISTANT (AZG) transporters, equilibrative nucleoside transporters (ENT), and Sugars Will Eventually be Exported Transporters (SWEET) have been characterized as involved in cytokinin transport processes. This review begins by introducing the spatial distributions of various cytokinins and the subcellular localizations of the proteins involved in cytokinin metabolism and signaling. Highlights focus on an inventory of the characterized transporters involved in cytokinin compartmentalization, including long-distance, intercellular, and intracellular transport, and the regulation of spatial distributions of cytokinins by environmental cues. Future directions for cytokinin research are also discussed."
Authors: Yanyan Tang, Zhong Huang, Shaohui Xu, Wenjie Zhou, Jianjun Ren, Fuxin Yu, Jingshan Wang, Wujun Ma and Lixian Qiao.
The Crop Journal (2024)
Abstract: "Ethylene plays essential roles in plant growth, development and stress responses. The ethylene signaling pathway and molecular mechanism have been studied extensively in Arabidopsis and rice but limited in peanuts. Here, we established a sand-culture method to screen pingyangmycin mutagenized peanut lines based on their specific response to ethylene (“triple response“). An ethylene-insensitive mutant, inhibition of peanut hypocotyl elongation 1 (iph1), was identified that showed reduced sensitivity to ethylene in both hypocotyl elongation and root growth. Through bulked segregant analysis sequencing, a major gene related to iph1, named AhIPH1, was preliminarily mapped at the chromosome Arahy.01, and further narrowed to a 450-kb genomic region through substitution mapping strategy. A total of 7014 genes were differentially expressed among the ACC treatment through RNA-seq analysis, of which only the Arahy.5BLU0Q gene in the candidate mapping interval was differentially expressed between WT and mutant iph1. Integrating sequence variations, functional annotation and transcriptome analysis revealed that a predicated gene, Arahy.5BLU0Q, encoding SNF1 protein kinase, may be the candidate gene for AhIPH1. This gene contained two single-nucleotide polymorphisms at promoter region and was more highly expressed in iph1 than WT. Our findings reveal a novel ethylene-responsive gene, which provides a theoretical foundation and new genetic resources for the mechanism of ethylene signaling in peanuts"
Authors: María Isabel Puga, César Poza-Carrión, Iris Martinez-Hevia, Laura Perez-Liens and Javier Paz-Ares
Journal of Plant Research (2024)
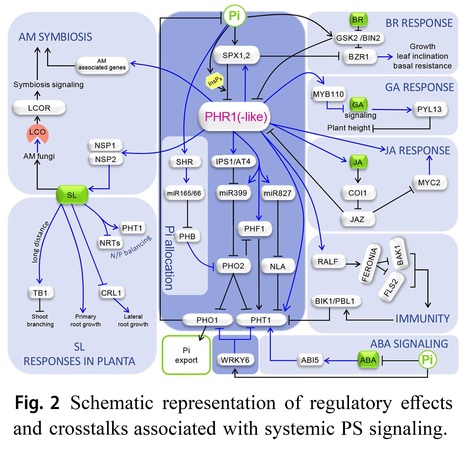
Abstract: "Phosphorus is indispensable for plant growth and development, with its status crucial for determining crop productivity. Plants have evolved various biochemical, morphological, and developmental responses to thrive under conditions of low P availability, as inorganic phosphate (Pi), the primary form of P uptake, is often insoluble in soils. Over the past 25 years, extensive research has focused on understanding these responses, collectively forming the Pi starvation response system. This effort has not only expanded our knowledge of strategies to cope with Pi starvation (PS) but also confirmed their adaptive significance. Moreover, it has identified and characterized numerous components of the intricate regulatory network governing P homeostasis. This review emphasizes recent advances in PS signaling, particularly highlighting the physiological importance of local PS signaling in inhibiting primary root growth and uncovering the role of TORC1 signaling in this process. Additionally, advancements in understanding shoot-root Pi allocation and a novel technique for studying Pi distribution in plants are discussed. Furthermore, emerging data on the regulation of plant-microorganism interactions by the PS regulatory system, crosstalk between the signaling pathways of phosphate starvation, phytohormones and immunity, and recent studies on natural variation in Pi homeostasis are addressed.
|


 Your new post is loading...
Your new post is loading...