Your new post is loading...
 Your new post is loading...
Authors: Jingyi Han (撖静宜), Thomas Welch, Ute Voß, Teva Vernoux, Rahul Bhosale and Anthony Bishopp.
iScience (2024)
Highlights • The first intron of ARF7 is required for expression in the root apical meristem. • It is not required for expression in either lateral roots or the shoot apex. • We find no binding sites in the first intron required for expression in root tips. • We propose that this intron exerts its effects via Intron Mediated Enhancement.
Abstract: "Auxin regulates plant growth and development through the transcription factors of the AUXIN RESPONSE FACTOR (ARF) gene family. ARF7 is one of five activators that bind DNA and elicit downstream transcriptional responses. In roots, ARF7 regulates growth, gravitropism and redundantly with ARF19, lateral root organogenesis. In this study we analyzed ARF7 cis-regulation, using different non-coding sequences of the ARF7 locus to drive GFP. We show that constructs containing the first intron led to increased signal in the root tip. Although bioinformatics analyses predicted several transcription factor binding sites in the first intron, we were unable to significantly alter expression of GFP in the root by mutating these. We instead observed the intronic sequences needed to be present within the transcribed sequences to drive expression in the root meristem. These data support a mechanism by which Intron Mediated Enhancement regulates the tissue specific expression of ARF7 in the root meristem."
Authors: Tomás Urzúa Lehuedé, Victoria Berdion Gabarain, Tomas Moyano, Lucia Ferrero, Miguel Angel Ibeas, Hernan Salinas-Grenet, Gerardo Núñez-Lillo, Romina Acha, Jorge Perez, Florencia Perotti, Virginia Natali Miguel, Fiorella Paola Spies, Miguel A. Rosas, Michitaro Shibata, Diana R. Rodríguez-García, Adrian A. Moreno, Keiko Sugimoto, Karen Sanguinet, Claudio Meneses, Raquel L. Chan, Federico Ariel, Jose M. Alvarez and José M. Estevez.
bioRxiv (2024)
Abstract: "The root hair (RH) cells can elongate to several hundred times their initial size, and are an ideal model system for investigating cell size control. Their development is influenced by both endogenous and external signals, which are combined to form a integrative response. Surprisingly, a low temperature condition of 10°C causes an increased RH growth in Arabidopsis and in several monocots, even when the development of the rest of the root and aerial parts of the plant are halted. Previously, we demonstrated a strong correlation between the growth response and a significant decrease in nutrient availability in the medium under low temperature conditions. However, the molecular basis responsible for receiving and transmitting signals related to the availability of nutrients in the soil, and their relation to plant development, remain largely unknown. We decided to further investigate the intricate molecular processes behind the particular responsiveness of this root cell type at low temperature. In this study, we have discovered a gene regulatory network (GRN) controlling early transcriptome responses to low temperature. This GNR is commanded by specific transcription factors (FTs), namely ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), a member of the homeodomain leucine zipper (HD-Zip I) group I 13 (AtHB13), the trihelix TF GT2-LIKE1 (GTL1), and a previously unidentified MYB-like TF (AT2G01060). Furthermore, we have identified four downstream RSL4 targets AtHB16, AtHB23, EARLY-RESPONSIVE TO DEHYDRATION 7 (ERD7) and ERD10 suggesting their participation in the regulation of RH development under these conditions. Functional analysis shows that such components of the RSL4-dependent transcriptional cascade influence the subsequent RH growth response to low temperature. These discoveries enhance our comprehension of how plants synchronize the RH growth in response to variations in temperature and nutrient availability at the cellular level."
Authors: Shammi Akter, Oscar Castaneda-Méndez and Jesús Beltrán
Current Opinion in Biotechnology (2024)
Highlights • Several challenges remain for the widespread use of synthetic biology in crops. • Synthetic gene expression circuitry is advancing in model plants. • Rapid enzyme discovery and testing are needed for plant metabolic engineering.
Abstract: "Plant synthetic biology (Plant SynBio) is an emerging field with the potential to enhance agriculture, human health, and sustainability. Integrating genetic tools and engineering principles, Plant SynBio aims to manipulate cellular functions and construct novel biochemical pathways to develop plants with new phenotypic traits, enhanced yield, and be able to produce natural products and pharmaceuticals. This review compiles research efforts in reprogramming plant developmental and biochemical pathways. We highlight studies leveraging new gene expression toolkits to alter plant architecture for improved performance in model and crop systems and to produce useful metabolites in plant tissues. Furthermore, we provide insights into the challenges and opportunities associated with the adoption of Plant SynBio in addressing complex issues impacting agriculture and human health."
Authors: Ping Yun, Cengiz Kaya and Sergey Shabala.
The Crop Journal (2024)
Abstract: "Salinity stress is a major environmental stress affecting crop productivity, and its negative impact on global food security is only going to increase, due to current climate trends. Salinity tolerance was present in wild crop relatives but significantly weakened during domestication. Regaining it back requires a good understanding of molecular mechanisms and traits involved in control of plant ionic and ROS homeostasis. This review summarizes our current knowledge on the role of major plant hormones (auxin, cytokinins, abscisic acid, salicylic acid, and jasmonate) in plants adaptation to soil salinity. We firstly discuss the role of hormones in controlling root tropisms, root growth and architecture (primary root elongation, meristematic activity, lateral root development, and root hairs formation). Hormone-mediated control of uptake and sequestration of key inorganic ions (sodium, potassium, and calcium) is then discussed followed by regulation of cell redox balance and ROS signaling in salt-stressed roots. Finally, the role of epigenetic alterations such as DNA methylation and histone modifications in control of plant ion and ROS homeostasis and signaling is discussed. This data may help develop novel strategies for breeding and cultivating salt-tolerant crops and improving agricultural productivity in saline regions."
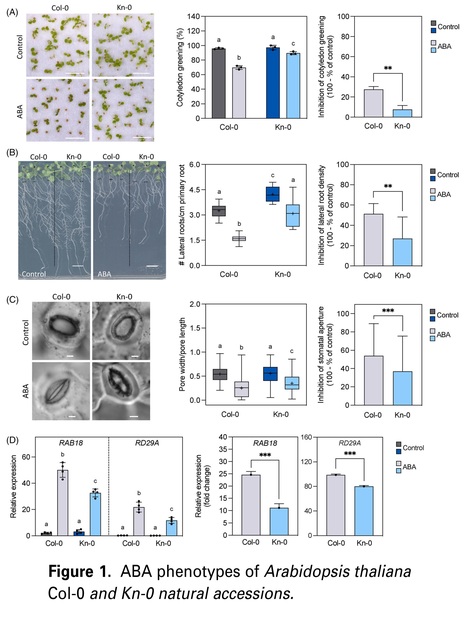
Authors: Alba R. Díez, Dóra Szakonyi, Jorge Lozano-Juste and Paula Duque.
The Plant Journal (2024)
Significance Statement: A comparative transcriptomic analysis of two Arabidopsis natural variants with distinct abscisic acid (ABA) sensitivity indicates that alternative splicing may contribute more than gene expression changes to the differential ABA response of the ecotypes. Our results support the notion of alternative splicing as a key regulatory layer of ABA-mediated stress responses.
Abstract: "Abscisic acid (ABA) is a crucial player in plant responses to the environment. It accumulates under stress, activating downstream signaling to implement molecular responses that restore homeostasis. Natural variance in ABA sensitivity remains barely understood, and the ABA pathway has been mainly studied at the transcriptional level, despite evidence that posttranscriptional regulation, namely, via alternative splicing, contributes to plant stress tolerance. Here, we identified the Arabidopsis accession Kn-0 as less sensitive to ABA than the reference Col-0, as shown by reduced effects of the hormone on seedling establishment, root branching, and stomatal closure, as well as by decreased induction of ABA marker genes. An in-depth comparative transcriptome analysis of the ABA response in the two variants revealed lower expression changes and fewer genes affected for the least ABA-sensitive ecotype. Notably, Kn-0 exhibited reduced levels of the ABA-signaling SnRK2 protein kinases and lower basal expression of ABA-reactivation genes, consistent with our finding that Kn-0 contains less endogenous ABA than Col-0. ABA also markedly affected alternative splicing, primarily intron retention, with Kn-0 being less responsive regarding both the number and magnitude of alternative splicing events, particularly exon skipping. We find that alternative splicing introduces a more ecotype-specific layer of ABA regulation and identify ABA-responsive splicing changes in key ABA pathway regulators that provide a functional and mechanistic link to the differential sensitivity of the two ecotypes. Our results offer new insight into the natural variation of ABA responses and corroborate a key role for alternative splicing in implementing ABA-mediated stress responses."
Authors: Emese Eysholdt-Derzsó, Bettina Hause, Margret Sauter and Romy R. Schmidt-Schippers.
Plant, Cell & Environment (2024)
Summary Statement: ERFVII pathway actively maintains abscisic acid homoeostasis, enabling a partial control of lateral root development under hypoxic conditions.
Abstract: "Oxygen limitation (hypoxia), arising as a key stress factor due to flooding, negatively affects plant development. Consequently, maintaining root growth under such stress is crucial for plant survival, yet we know little about the root system's adaptions to low-oxygen conditions and its regulation by phytohormones. In this study, we examine the impact of hypoxia and, herein, the regulatory role of group VII ETHYLENE-RESPONSE FACTOR (ERFVII) transcription factors on root growth in Arabidopsis. We found lateral root (LR) elongation to be actively maintained by hypoxia via ERFVII factors, as erfVII seedlings possess hypersensitivity towards hypoxia regarding their LR growth. Pharmacological inhibition of abscisic acid (ABA) biosynthesis revealed ERFVII-driven counteraction of hypoxia-induced inhibition of LR formation in an ABA-dependent manner. However, postemergence LR growth under hypoxia mediated by ERFVIIs was independent of ABA. In roots, ERFVIIs mediate, among others, the induction of ABA-degrading ABA 8′-hydroxylases CYP707A1 expression. RAP2.12 could activate the pCYC707A1:LUC reporter gene, indicating, combined with single mutant analyses, that this transcription factor regulates ABA levels through corresponding transcript upregulation. Collectively, hypoxia-induced adaptation of the Arabidopsis root system is shaped by developmental reprogramming, whereby ERFVII-dependent promotion of LR emergence, but not elongation, is partly executed through regulation of ABA degradation."
Authors: Renata P. Pincelli-Souza, Qian Tang, Brandon M. Miller and Jerry D. Cohen.
Horticultural Advances (2024)
Abstract: "The first reports that auxins promoted root formation in cuttings and that indole-3-butyric acid (IBA) was a particularly effective treatment date from the early 1930s. Since its introduction into horticultural practice, the focus on improvements in the rooting of plants has been largely on the proper use of auxins to enhance adventitious rooting (AR) as well as to increase the range of plants where it can be effective. In this review, we focus on new ideas that might build on what is known about auxin induction of AR. We explore what the evolution in chemical biology has opened through novel high-throughput screening tools to explore auxin regulation of plant development and what it might add to our understanding and potential to produce new tools for the manipulation of AR. The potential for using stronger auxin analogues, alternative indolealkanoic acids, compounds that alter β-oxidation of IBA and other indolealkanoic acids, auxin conjugates, inhibitors of auxin conjugation, inhibitors of endogenous auxin biosynthesis, as well as other plant hormones and compounds that inhibit the production or mimic the effects of signals that might be involved in AR are all discussed. The expectation is that a summary of these advances in our understanding of the chemical biology important to AR might increase the use and exploration of new ideas for the improvement in the practical approaches to advance horticultural rooting methods."
Author: Aida Maric.
Plant Physiology (2024)
Excerpts: "Recent work by Kong et al. (2024), published in Plant Physiology reports ethylene-auxin hormonal crosstalk as a regulator of root angle in cereal crops. The authors used different root phenotyping methods to compare root response of wild-type and ethylene-insensitive rice and maize mutants. EIL1 and EIN2 are crucial links in the ethylene signaling pathway, conducting ethylene signal from the endoplasmic reticulum to the nucleus to control ethylene-responsive genes. Rice and maize mutants of EIL1 and EIN2 (oseil1, osein2 and zmein2-1) were not able to sense ethylene signals and had impaired ethylene response. Kong et al. (2024) showed that oseil1 and osein2 mutants have a shallower root system with larger root angles (Figure 1A). Additionally, ethylene mutants had an impaired gravitropic response compared to wild-type plants."
"By comparing hormone profiles of ethylene-insensitive mutants with wild-type plants they showed that ethylene-insensitive mutants exhibit lower levels of auxin. Exogenous application of auxin could rescue the root angle phenotype of the ethylene-insensitive mutants (Figure 1B), pinpointing auxin as the intermediate signal in ethylene-mediated root growth angles. Furthermore the authors showed that MHZ10, a member of the auxin biosynthesis pathway, plays a key role in the ethylene-mediated regulation of root angle (Figure 1B)."
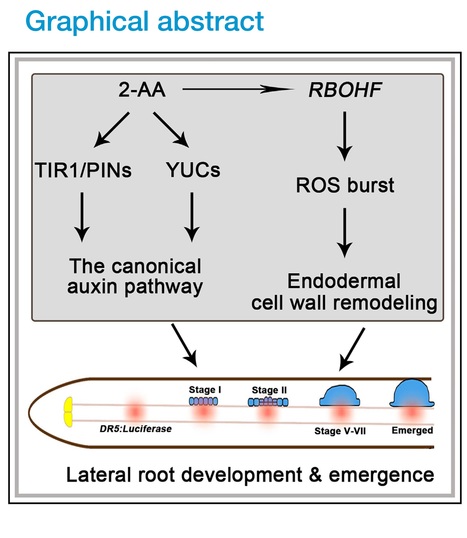
Authors: Yu Chen, Yansong Fu, Yanwei Xia, Youzhi Miao, Jiahui Shao, Wei Xuan, Yunpeng Liu, Weibing Xun, Qiuyan Yan, Qirong Shen and Ruifu Zhang.
Cell Reports (2023)
Editor's view: Chen et al. show that 2-AA, a compound identified from T. guizhouense NJAU4742, promotes plant lateral root development via the canonical auxin pathway as a stimulator and increases ROS deposition in the Casparian strip as an IAA mimic, which finally enhances endodermal cell wall remodeling and facilitates lateral root emergence.
Highlights: • 2-AA identified from T. guizhouense NJAU4742 can promote lateral root development • 2-AA regulates auxin signaling and transport in the canonical auxin pathway • 2-AA enhances endodermal cell wall remodeling via an RBOHF-induced ROS burst
Abstract: "Trichoderma spp. have evolved the capacity to communicate with plants by producing various secondary metabolites (SMs). Nonhormonal SMs play important roles in plant root development, while specific SMs from rhizosphere microbes and their underlying mechanisms to control plant root branching are still largely unknown. In this study, a compound, anthranilic acid (2-AA), is identified from T. guizhouense NJAU4742 to promote lateral root development. Further studies demonstrate that 2-AA positively regulates auxin signaling and transport in the canonical auxin pathway. 2-AA also partly rescues the lateral root numbers of CASP1pro:shy2-2, which regulates endodermal cell wall remodeling via an RBOHF-induced reactive oxygen species burst. In addition, our work reports another role for microbial 2-AA in the regulation of lateral root development, which is different from its better-known role in plant indole-3-acetic acid biosynthesis. In summary, this study identifies 2-AA from T. guizhouense NJAU4742, which plays versatile roles in regulating plant root development."
Author: Gwendolyn K. Kirschner.
The Plant Cell (2024)
Excerpts: "Yuxiang Li, Juan Wang and colleagues (Li et al. 2024) now connect the two roles of ethylene: as a signal responding to soil compaction and to trigger crown root development. In this work, they mimicked different levels of compaction by increasing agar concentrations and compared the growth of wildtype plants and ethylene signaling mutants. They found that compaction triggered crown root initiation via an ethylene-dependent pathway (Figure, A), and manipulating ethylene-related regulators influenced both root development and grain yield, implying that these regulators balance both root and grain development."
"By analyzing crown root numbers and transcript levels in oswox11 and ethylene signaling mutants, or overexpression combinations under normal or compacted soil conditions, the authors confirmed that the ethylene-OsEIL1-OsWOX11 module facilitates crown root development in compacted soil (Figure, B)."
Authors: Yuxiang Li, Juan Wang, Yadi Gao, Bipin K Pandey, Lucas León Peralta Ogorek, Yu Zhao, Ruidang Quan, Zihan Zhao, Lei Jiang, Rongfeng Huang and Hua Qin.
The Plant Cell (2024)
One-sentence summary: The phytohormone ethylene fine-tunes rice crown root development by activating WUSCHEL-RELATED HOMEOBOX 11 expression in response to soil compaction.
Abstract: "Optimizing the root architecture of crops is an effective strategy for improving crop yields. Soil compaction is a serious global problem that limits crop productivity by restricting root growth, but the underlying molecular mechanisms are largely unclear. Here, we show that ethylene stimulates rice (Oryza sativa) crown root development in response to soil compaction. First, we demonstrate that compacted soil promotes ethylene production and the accumulation of ETHYLENE INSENSITIVE 3-LIKE 1 (OsEIL1) in rice roots, stimulating crown root primordia initiation and development, thereby increasing crown root number in lower stem nodes. Through transcriptome profiling and molecular analyses, we reveal that OsEIL1 directly activates the expression of WUSCHEL-RELATED HOMEOBOX 11 (OsWOX11), an activator of crown root emergence and growth, and that OsWOX11 mutations delay crown root development, thus impairing the plant’s response to ethylene and soil compaction. Genetic analysis demonstrates that OsWOX11 functions downstream of OsEIL1. In summary, our results demonstrate that the OsEIL1–OsWOX11 module regulates ethylene action during crown root development in response to soil compaction, providing a strategy for the genetic modification of crop root architecture and grain agronomic traits."
Authors: Zipeng Yu, Xingzhen Qu, Bingsheng Lv, Xiaoxuan Li, Jiaxuan Sui, Qianqian Yu and Zhaojun Ding.
The Plant Cell (2024)
One-sentence summary: Auxin removes the barrier protein ERF13 through the MPK14-MAC3A and MAC3B signaling module, thus promoting lateral root emergence.
Abstract: "Lateral roots (LRs) increase root surface area and allow plants greater access to soil water and nutrients. LR formation is tightly regulated by the phytohormone auxin. Whereas the transcription factor ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR13 (ERF13) prevents LR emergence in Arabidopsis (Arabidopsis thaliana), auxin activates MITOGEN-ACTIVATED PROTEIN KINASE14 (MPK14), which leads to ERF13 degradation and ultimately promotes LR emergence. In this study, we discovered interactions between ERF13 and the E3 ubiquitin ligases MOS4-ASSOCIATED COMPLEX 3A (MAC3A) and MAC3B. As MAC3A and MAC3B gradually accumulate in the LR primordium, ERF13 levels gradually decrease. We demonstrate that MAC3A and MAC3B ubiquitinate ERF13, leading to its degradation and accelerating the transition of LR primordia from stages IV to V. Auxin enhances the MAC3A and MAC3B interaction with ERF13 by facilitating MPK14-mediated ERF13 phosphorylation. In summary, this study reveals the molecular mechanism by which auxin eliminates the inhibitory factor ERF13 through the MPK14-MAC3A and MAC3B signaling module, thus promoting LR emergence."
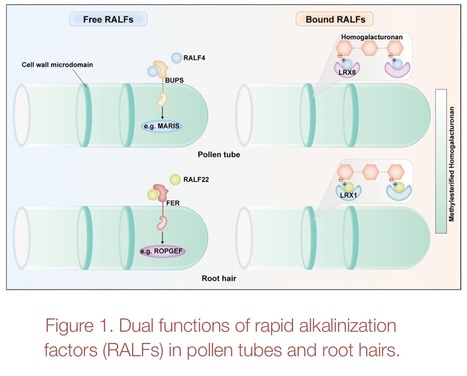
Authors: Jia Chen, Feng Yu and Fan Xu.
Trends in Plant Science (2024)
Abstract: "Rapid alkalinization factors (RALFs) have long been known to act as signaling molecules in plant cells, but whether they affect cell wall (CW) patterning and expansion remains unclear. Very recent advances in tip-growing cells showed that positively charged RALFs affect key attributes of the structural components of the nascent CW."
|
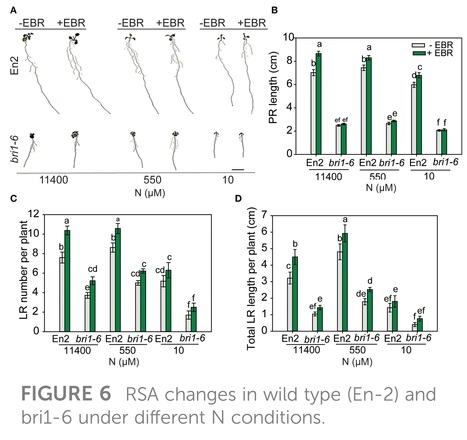
Authors: Mahamud Hossain Al-Mamun, Christopher Ian Cazzonelli and Priti Krishna.
Frontiers in Plant Science (2024)
Abstract: "Plants modify their root system architecture (RSA) in response to nitrogen (N) deficiency. The plant steroidal hormone, brassinosteroid (BR), plays important roles in root growth and development. This study demonstrates that optimal levels of exogenous BR impact significant increases in lateral root length and numbers in Arabidopsis seedlings under mild N-deficient conditions as compared to untreated seedlings. The impact of BR on RSA was stronger under mild N deficiency than under N-sufficient conditions. The BR effects on RSA were mimicked in dominant mutants of BZR1 and BES1 (bzr1-1D and bes1-D) transcription factors, while the RSA was highly reduced in the BR-insensitive mutant bri1-6, confirming that BR signaling is essential for the development of RSA under both N-sufficient and N-deficient conditions. Exogenous BR and constitutive activity of BZR1 and BES1 in dominant mutants led to enhanced root meristem, meristematic cell number, and cortical cell length. Under mild N deficiency, bzr1-1D displayed higher fresh and dry shoot weights, chlorophyll content, and N levels in the shoot, as compared to the wild type. These results indicate that BR modulates RSA under both N-sufficient and N-deficient conditions via the transcription factors BES1/BZR1 module and confers tolerance to N deficiency."
Authors: Mengdi Fu, Xiaolei Yao, Xiaolin Li, Jing Liu, Mengyan Bai, Zijun Fang, Jiming Gong, Yuefeng Guan and Fang Xie.
The Plant Journal (2024)
Significance Statement: GmNLP1 and GmNLP4 activate nitrate-induced CLE peptides NIC1a/b to mediate nitrate-regulated root nodulation.
Abstract: "Symbiotic nitrogen fixation is an energy-intensive process, to maintain the balance between growth and nitrogen fixation, high concentrations of nitrate inhibit root nodulation. However, the precise mechanism underlying the nitrate inhibition of nodulation in soybean remains elusive. In this study, CRISPR-Cas9-mediated knockout of GmNLP1 and GmNLP4 unveiled a notable nitrate-tolerant nodulation phenotype. GmNLP1b and GmNLP4a play a significant role in the nitrate-triggered inhibition of nodulation, as the expression of nitrate-responsive genes was largely suppressed in Gmnlp1b and Gmnlp4a mutants. Furthermore, we demonstrated that GmNLP1b and GmNLP4a can bind to the promoters of GmNIC1a and GmNIC1b and activate their expression. Manipulations targeting GmNIC1a and GmNIC1b through knockdown or overexpression strategies resulted in either increased or decreased nodule number in response to nitrate. Additionally, transgenic roots that constitutively express GmNIC1a or GmNIC1b rely on both NARK and hydroxyproline O-arabinosyltransferase RDN1 to prevent the inhibitory effects imposed by nitrate on nodulation. In conclusion, this study highlights the crucial role of the GmNLP1/4-GmNIC1a/b module in mediating high nitrate-induced inhibition of nodulation."
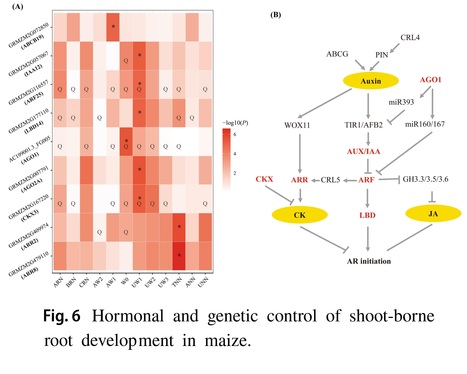
Authors: Pengcheng Li, Zhihai Zhang, Gui Xiao, Zheng Zhao, Kunhui He, Xiaohong Yang, Qingchun Pan, Guohua Mi, Zhongtao Jia, Jianbing Yan, Fanjun Chen and Lixing Yuan.
Theoretical and Applied Genetics (2024)
Abstract: "Efficient nutrient and water acquisition from soils depends on the root system architecture (RSA). However, the genetic determinants underlying RSA in maize remain largely unexplored. In this study, we conducted a comprehensive genetic analysis for 14 shoot-borne root traits using 513 inbred lines and 800 individuals from four recombinant inbred line (RIL) populations at the mature stage across multiple field trails. Our analysis revealed substantial phenotypic variation for these 14 root traits, with a total of 389 and 344 QTLs identified through genome-wide association analysis (GWAS) and linkage analysis, respectively. These QTLs collectively explained 32.2–65.0% and 23.7–63.4% of the trait variation within each population. Several a priori candidate genes involved in auxin and cytokinin signaling pathways, such as IAA26, ARF2, LBD37 and CKX3, were found to co-localize with these loci. In addition, a total of 69 transcription factors (TFs) from 27 TF families (MYB, NAC, bZIP, bHLH and WRKY) were found for shoot-borne root traits. A total of 19 genes including PIN3, LBD15, IAA32, IAA38 and ARR12 and 19 GWAS signals were overlapped with selective sweeps. Further, significant additive effects were found for root traits, and pyramiding the favorable alleles could enhance maize root development. These findings could contribute to understand the genetic basis of root development and evolution, and provided an important genetic resource for the genetic improvement of root traits in maize."
Via Jean-Michel Ané
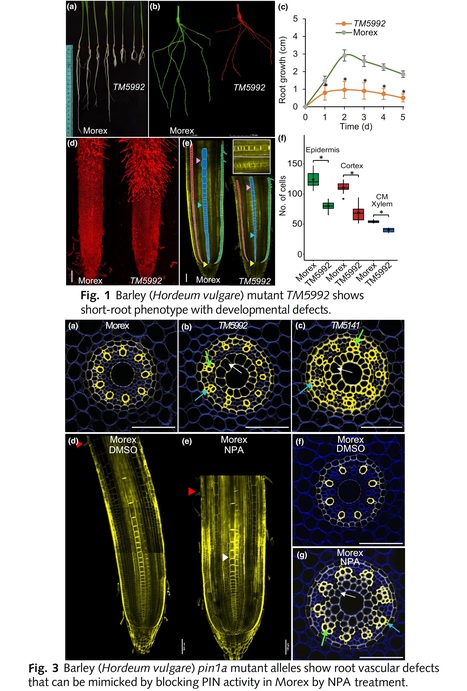
Authors: Riccardo Fusi, Sara Giulia Milner, Serena Rosignoli, Riccardo Bovina, Cristovão De Jesus Vieira Teixeira, Haoyu Lou, Brian S. Atkinson, Aditi N. Borkar, Larry M. York, Dylan H. Jones, Craig J. Sturrock, Nils Stein, Martin Mascher, Roberto Tuberosa, Devin O'Connor, Malcolm J. Bennett, Anthony Bishopp, Silvio Salvi and Rahul Bhosale.
New Phytologist (2024)
Abstract: "Barley (Hordeum vulgare) is an important global cereal crop and a model in genetic studies. Despite advances in characterising barley genomic resources, few mutant studies have identified genes controlling root architecture and anatomy, which plays a critical role in capturing soil resources. Our phenotypic screening of a TILLING mutant collection identified line TM5992 exhibiting a short-root phenotype compared with wild-type (WT) Morex background. Outcrossing TM5992 with barley variety Proctor and subsequent SNP array-based bulk segregant analysis, fine mapped the mutation to a cM scale. Exome sequencing pinpointed a mutation in the candidate gene HvPIN1a, further confirming this by analysing independent mutant alleles. Detailed analysis of root growth and anatomy in Hvpin1a mutant alleles exhibited a slower growth rate, shorter apical meristem and striking vascular patterning defects compared to WT. Expression and mutant analyses of PIN1 members in the closely related cereal brachypodium (Brachypodium distachyon) revealed that BdPIN1a and BdPIN1b were redundantly expressed in root vascular tissues but only Bdpin1a mutant allele displayed root vascular defects similar to Hvpin1a. We conclude that barley PIN1 genes have sub-functionalised in cereals, compared to Arabidopsis (Arabidopsis thaliana), where PIN1a sequences control root vascular patterning."
Authors: Xiuzhen Kong, Suhang Yu, Yali Xiong, Xiaoyun Song, Lucia Nevescanin-Moreno, Xiaoqing Wei, Jinliang Rao, Hu Zhou, Malcolm J. Bennett, Bipin K. Pandey and Guoqiang Huang.
Current Biology (2024)
Editor's view: Kong et al. provide evidence for the anchorage functions of root hairs in root penetration, a process that is regulated by OsYUC8-mediated auxin biosynthesis and OsAUX1-mediated auxin transport when roots encounter a compacted barrier.
Highlights • Mechanical impedance induced the upregulation of auxin biosynthesis in root apex • Auxin transport from root tip to hair zone promoted root penetration via root hair • Mutants of OsYUC8 and OsAUX1 had trouble penetrating into the compacted layer • Increased root hair length provided greater anchorage force for root penetration
Abstract: "Compacted soil layers adversely affect rooting depth and access to deeper nutrient and water resources, thereby impacting climate resilience of crop production and global food security. Root hair plays well-known roles in facilitating water and nutrient acquisition. Here, we report that root hair also contributes to root penetration into compacted layers. We demonstrate that longer root hair, induced by elevated auxin response during a root compaction response, improves the ability of rice roots to penetrate harder layers. This compaction-induced auxin response in the root hair zone is dependent on the root apex-expressed auxin synthesis gene OsYUCCA8 (OsYUC8), which is induced by compaction stress. This auxin source for root hair elongation relies on the auxin influx carrier AUXIN RESISTANT 1 (OsAUX1), mobilizing this signal from the root apex to the root hair zone. Mutants disrupting OsYUC8 and OsAUX1 genes exhibit shorter root hairs and weaker penetration ability into harder layers compared with wild type (WT). Root-hair-specific mutants phenocopy these auxin-signaling mutants, as they also exhibit an attenuated root penetration ability. We conclude that compaction stress upregulates OsYUC8-mediated auxin biosynthesis in the root apex, which is subsequently mobilized to the root hair zone by OsAUX1, where auxin promotes root hair elongation, improving anchorage of root tips to their surrounding soil environment and aiding their penetration ability into harder layers."
Authors: Yuwen Zhang, Xingliang Duan, Zhen Wang, Yuanda Lv, Weicong Qi, Lun Li, Le Luo and Wei Xuan.
Biochemical and Biophysical Research Communications (2024)
Highlights • Both synthetic AtCEPs and AtCEP5 overexpression suppress primary root elongation and lateral root formation. • The regulation of CEPs on root development requires their receptors CEPRs. • Molecular evidence reveals that CEPs inhibit root development by modulating auxin and cytokinin signaling.
Abstract: "C-terminally encoded peptides (CEPs) are peptide hormones that function as mobile signals coordinating crucial developmental programs in plants. Previous studies have revealed that CEPs exert negative regulation on root development through interaction with CEP receptors (CEPRs), CEPR DOWNSTREAMs (CEPDs), the cytokinin receptor ARABIDOPSIS HISTIDINE KINASE (AHKs) and the transcriptional repressor Auxin/Indole-3-Acetic Acid (AUX/IAA). However, the precise molecular mechanisms underlying CEPs-mediated regulation of root development via auxin and cytokinin signaling pathways still necessitate further detailed investigation. In this study, we examined prior research and elucidated the underlying molecular mechanisms. The results showed that both synthetic AtCEPs and overexpression of AtCEP5 markedly suppressed primary root elongation and lateral root (LR) formation in Arabidopsis. Molecular biology and genetics elucidated how CEPs inhibit root growth by suppressing auxin signaling while promoting cytokinin signaling. In summary, this study elucidated the inhibitory effects of AtCEPs on Arabidopsis root growth and provided insights into their potential molecular mechanisms, thus enhancing our comprehension of CEP-mediated regulation of plant growth and development."
Authors: Clare Hurst and Miriam L. Gifford.
Development (2024)
Excepts: "Legumes form a symbiotic relationship with bacteria called rhizobia. The rhizobia fix atmospheric dinitrogen, which is used by the plant as a nitrogen resource, and take up molecules, including carbon compounds, from the plant. Rhizobia are housed in nodules that develop on roots after a series of orchestrated stages that are triggered by the perception and entry of rhizobia at root hairs (reviewed by Luo et al., 2023). Study of this relationship and the mechanisms that govern nodule formation and inhibition may reveal targets to promote enhanced nodulation efficiency and increased nitrogen acquisition for the plant. Phytohormones have been shown to play a key role in both nodule formation and inhibition; therefore, a better understanding of hormonal regulatory activity could provide options for such enhancement. To address this, Drapek and colleagues (2023 preprint) focus on the dynamics of gibberellin (GA), a phytohormone previously identified as both a positive and negative regulator of nodulation (Rizza et al., 2017; Fonouni-Farde et al., 2016).
"In conclusion, this study analysed the spatial and temporal accumulation of GA, the interactions between GA and key nodule identity regulatory genes, and tested the importance of GA biosynthesis enzymes in nodule formation. The work provides strong evidence that GA is a potential determinator between lateral root formation and nodulation, and helps to explain previous contradictory findings related to the importance of GA, as well as cytokinin, in these processes."
Authors: Zachary Aldiss, Hannah Robinson, Yasmine Lam, Richard Dixon, Peter A. Crisp, Ian D. Godwin, Andrew Borrell, Lee Hickey and Karen Massel.
bioRxiv (2024)
Abstract: "Roots provide the critical interface where plants acquire nutrients and water, but our limited understanding of the genetic controls modulating root system architecture (RSA) in crop species constrains opportunities to develop future cultivars with improved root systems. However, there is vast knowledge of root developmental genes in model plant species, which has the potential to accelerate progress in crops with more complex genomes, particularly given that genome editing protocols are now available for most species. PIN-FORMED2 (PIN2) encodes a root specific polar auxin transporter, where its absence resulted in roots being unable to orient themselves using gravity, producing a significantly wider root system. To explore the role of PIN2 in a cereal crop, we used CRISPR/Cas9 editing to knockout of PIN2 in barley (Hordeum vulgare). Like Arabidopsis, the roots of barley pin2 loss-of-function mutants displayed an agravitropic response at seedling growth stages, resulting in a significantly shallower and wider root system at later growth stages. Notably, despite the significant change in RSA, there was no change in shoot architecture or total shoot biomass. We discuss the future challenges and opportunities to harness the PIN2 pathway to optimise RSA in crops for a range of production scenarios without a shoot trade-off."
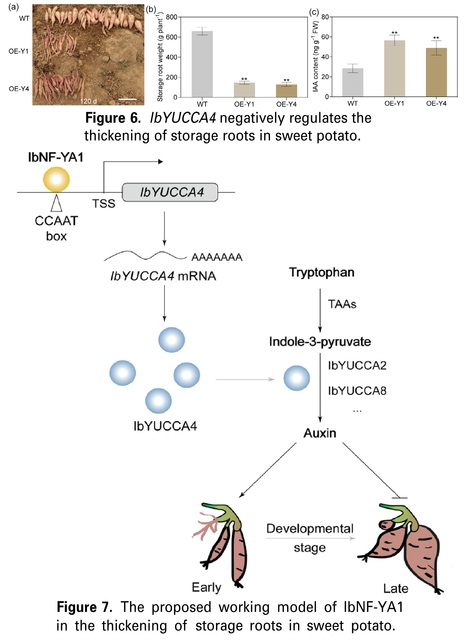
Authors: Luyao Xue, Yuxin Wang, Yue Fan, Zhicheng Jiang, Zihao Wei, Hong Zhai, Shaozhen He, Huan Zhang, Yufeng Yang, Ning Zhao, Shaopei Gao and Qingchang Liu.
The Plant Journal (2024)
Abstract: "As a major worldwide root crop, the mechanism underlying storage root yield formation has always been a hot topic in sweet potato [Ipomoea batatas (L.) Lam.]. Previously, we conducted the transcriptome database of differentially expressed genes between the cultivated sweet potato cultivar “Xushu18,” its diploid wild relative Ipomoea triloba without storage root, and their interspecific somatic hybrid XT1 with medium-sized storage root. We selected one of these candidate genes, IbNF-YA1, for subsequent analysis. IbNF-YA1 encodes a nuclear transcription factor Y subunit alpha (NF-YA) gene, which is significantly induced by the natural auxin indole-3-acetic acid (IAA). The storage root yield of the IbNF-YA1 overexpression (OE) plant decreased by 29.15–40.22% compared with the wild type, while that of the RNAi plant increased by 10.16–21.58%. Additionally, IAA content increased significantly in OE plants. Conversely, the content of IAA decreased significantly in RNAi plants. Furthermore, real-time quantitative reverse transcription-PCR (qRT-PCR) analysis demonstrated that the expressions of the key genes IbYUCCA2, IbYUCCA4, and IbYUCCA8 in the IAA biosynthetic pathway were significantly changed in transgenic plants. The results indicated that IbNF-YA1 could directly target IbYUCCA4 and activate IbYUCCA4 transcription. The IAA content of IbYUCCA4 OE plants increased by 71.77–98.31%. Correspondingly, the storage root yield of the IbYUCCA4 OE plant decreased by 77.91–80.52%. These findings indicate that downregulating the IbNF-YA1 gene could improve the storage root yield in sweet potato."
Authors: Vijay Pratap Singh, Saumya Jaiswal, Yuanyuan Wang, Shouli Feng, Durgesh Kumar Tripathi, Samiksha Singh, Ravi Gupta, Dawei Xue, Shengchun Xu and Zhong-Hua Chen.
Trends in Plant Science (2024)
Highlights: Reactive oxygen species (ROS) are considered toxic substances causing oxidative damage in plants, but they are also important signaling molecules crucial for many developmental processes of root and leaf. The origin of many major ROS signaling target proteins is highly conserved, predating the evolution of land plants. ROS function for the regulation of root and leaf development with the presence of phytohormones, gasotransmitters, and target proteins, particularly mitogen-activated protein kinases (MAPKs).
Abstract: "Reactive oxygen species (ROS) are the key players in regulating developmental processes of plants. Plants have evolved a large array of gene families to facilitate the ROS-regulated developmental process in roots and leaves. However, the cellular targets of ROS during plant evolutionary development are still elusive. Here, we found early evolution and large expansions of protein families such as mitogen-activated protein kinases (MAPK) in the evolutionarily important plant lineages. We review the recent advances in interactions among ROS, phytohormones, gasotransmitters, and protein kinases. We propose that these signaling molecules act in concert to maintain cellular ROS homeostasis in developmental processes of root and leaf to ensure the fine-tuning of plant growth for better adaptation to the changing climate."
Author: Rory Osborne.
The Plant Cell (2024)
Excerpts: " In this issue of The Plant Cell, Zipeng Yu, Xingzhen Qu and colleagues (Yu et al. 2024) beautifully characterize the mechanism underpinning the auxin-dependent posttranslational regulation of ETHYLENE RESPONSE FACTOR 13 (ERF13) (see Figure)."
"In this study, they explore how this degradation is regulated. In doing so, they reveal a novel role for MOS4-ASSOCIATED COMPLEX 3A (MAC3A) and 3B (MAC3B), so far only characterized as regulators of the plant immune response (Monaghan et al. 2009), in promoting the emergence of LRs."
"Thus, the authors conclude that auxin regulates ERF13 via two mechanisms: first, it activates MAPK14, which then phosphorylates ERF13, increasing its binding affinity to MAC3A/B, and secondly, it promotes expression of MAC3A/B at sites of LR initiation."
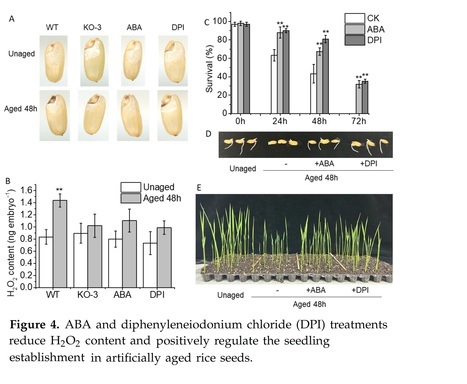
Authors: Qin Zheng, Zhenning Teng, Jianhua Zhang and Nenghui Ye.
Plants (2024)
Abstract: "The seed, a critical organ in higher plants, serves as a primary determinant of agricultural productivity, with its quality directly influencing crop yield. Improper storage conditions can diminish seed vigor, adversely affecting seed germination and seedling establishment. Therefore, understanding the seed-aging process and exploring strategies to enhance seed-aging resistance are paramount. In this study, we observed that seed aging during storage leads to a decline in seed vigor and can coincide with the accumulation of hydrogen peroxide (H2O2) in the radicle, resulting in compromised or uneven germination and asynchronous seedling emergence. We identified the abscisic acid (ABA) catabolism gene, abscisic acid 8′-hydroxylase 2 (OsABA8ox2), as significantly induced by aging treatment. Interestingly, transgenic seeds overexpressing OsABA8ox2 exhibited reduced seed vigor, while gene knockout enhanced seed vigor, suggesting its role as a negative regulator. Similarly, seeds pretreated with ABA or diphenyleneiodonium chloride (DPI, an H2O2 inhibitor) showed increased resistance to aging, with more robust early seedling establishment. Both OsABA8ox2 mutant seeds and seeds pretreated with ABA or DPI displayed lower H2O2 content during aging treatment. Overall, our findings indicate that ABA mitigates rice seed aging by reducing H2O2 accumulation in the radicle. This study offers valuable germplasm resources and presents a novel approach to enhancing seed resistance against aging."
|



 Your new post is loading...
Your new post is loading...