Your new post is loading...
 Your new post is loading...
Authors: Sonia Gazzarrini and Liang Song.
Annual Review of Plant Biology (2024)
Abstract: "Development is a chain reaction in which one event leads to another until the completion of a life cycle. Phase transitions are milestone events in the cycle of life. LEAFY COTYLEDON1 (LEC1), ABA INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 proteins, collectively known as LAFL, are master transcription factors (TFs) regulating seed and other developmental processes. Since the initial characterization of the LAFL genes (44, 71, 98–100), more than three decades of active research has generated tremendous amounts of knowledge about these TFs, whose roles in seed development and germination have been comprehensively reviewed (63, 43, 65, 81). Recent advances in cell biology and genetic and genomic tools have allowed the characterization of the LAFL regulatory networks in previously challenging tissues at a higher throughput and resolution in reference species and crops. In this review, we provide a holistic perspective by integrating advances at the epigenetic, transcriptional, posttranscriptional, and protein levels to exemplify the spatiotemporal regulation of the LAFL networks in Arabidopsis seed development and phase transitions, and we briefly discuss the evolution of these TF networks."
Authors: Hongjiao Jiang, Lijun Xie, Zhiqun Gu, Hongyao Mei, Haohao Wang, Jing Zhang, Minmin Wang, Yiteng Xu, Chuanen Zhou and Lu Han.
The Plant Journal (2024)
Significance Statement: Auxins play important roles in various aspects of plant growth and development. In Medicago truncatula, orthologs of PIN1-mediated auxin transportation are involved in various developmental processes and amino acid biosynthesis and metabolism in seeds, which offers insights into the molecular mechanisms governing seed size regulation in crops.
Abstract: "The regulation of seed development is critical for determining crop yield. Auxins are vital phytohormones that play roles in various aspects of plant growth and development. However, its role in amino acid biosynthesis and metabolism in seeds is not fully understood. In this study, we identified a mutant with small seeds through forward genetic screening in Medicago truncatula. The mutated gene encodes MtPIN4, an ortholog of PIN1. Using molecular approaches and integrative omics analyses, we discovered that auxin and amino acid content significantly decreased in mtpin4 seeds, highlighting the role of MtPIN4-mediated auxin distribution in amino acid biosynthesis and metabolism. Furthermore, genetic analysis revealed that the three orthologs of PIN1 have specific and overlapping functions in various developmental processes in M. truncatula. Our findings emphasize the significance of MtPIN4 in seed development and offer insights into the molecular mechanisms governing the regulation of seed size in crops. This knowledge could be applied to enhance crop quality by targeted manipulation of seed protein regulatory pathways."
Authors: R. B. Lima and D. D. Figueiredo.
Plant and Cell Physiology (2024)
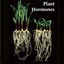
Abstract: "Since the discovery of brassinolide in the pollen of rapeseed, brassinosteroids (BRs) have consistently been associated with reproductive traits. However, compared to what is known for how BRs shape vegetative development, the understanding of how these hormones regulate reproductive traits is comparatively still lacking. Nevertheless, there is now considerable evidence that BRs regulate almost all aspects of reproduction, from ovule and pollen formation to seed and fruit development. Here, we review the current body of knowledge on how BRs regulate reproductive processes in plants, and what is known about how these pathways are transduced at the molecular level. We then discuss how the manipulation of BR biosynthesis and signaling can be a promising avenue for improving crop traits which rely on efficient reproduction. We thus propose that BR hold an untapped potential for plant breeding, which could contribute to attain food security in the coming years."
Authors: Avinash Sharma, Shalev Goldfarb, Dina Raveh and Dudy Bar-Zvi.
bioRxiv (2024)
Abstract: "Seed germination is a highly controlled developmental process. Prevention of early germination is essential to avoid germination at a time that is not likely to allow the completion of the plant life cycle before entering unfavorable seasons. The plant hormone ABA is the key regulator of seed dormancy and inhibition of germination. ABA is also involved in the plant response to drought. Here we report on the involvement of AtPUB41, a U-BOX E3 ubiquitin ligase from Arabidopsis thaliana, in regulating ABA signaling, seed dormancy, germination, and drought resilience. AtPUB41 is expressed in most plant vegetative and reproductive tissues. AtPUB41 protein is localized in the cytosol and nucleus. pub41 T-DNA insertion mutants display reduced seed dormancy, and their germination is less inhibited by exogenous ABA than seeds of wild-type plants. pub41 mutant plants are also hypersensitive to drought. ABA induces AtPUB41 promoter activity and steady-state mRNA levels in the roots. Our data suggest that AtPUB41 is a positive regulator of ABA signaling."
Authors: Tatsuki Akabane, Nobuhiro Suzuki, Kazuyoshi Ikeda, Tomoki Yonezawa, Satoru Nagatoishi, Hiroyoshi Matsumura, Takuya Yoshizawa, Wataru Tsuchiya, Satoshi Kamino, Kouhei Tsumoto, Ken Ishimaru, Etsuko Katoh and Naoki Hirotsu.
Scientific Reports (2024)
Abstract: "An indole-3-acetic acid (IAA)-glucose hydrolase, THOUSAND-GRAIN WEIGHT 6 (TGW6), negatively regulates the grain weight in rice. TGW6 has been used as a target for breeding increased rice yield. Moreover, the activity of TGW6 has been thought to involve auxin homeostasis, yet the details of this putative TGW6 activity remain unclear. Here, we show the three-dimensional structure and substrate preference of TGW6 using X-ray crystallography, thermal shift assays and fluorine nuclear magnetic resonance (19F NMR). The crystal structure of TGW6 was determined at 2.6 Å resolution and exhibited a six-bladed β-propeller structure. Thermal shift assays revealed that TGW6 preferably interacted with indole compounds among the tested substrates, enzyme products and their analogs. Further analysis using 19F NMR with 1,134 fluorinated fragments emphasized the importance of indole fragments in recognition by TGW6. Finally, docking simulation analyses of the substrate and related fragments in the presence of TGW6 supported the interaction specificity for indole compounds. Herein, we describe the structure and substrate preference of TGW6 for interacting with indole fragments during substrate recognition. Uncovering the molecular details of TGW6 activity will stimulate the use of this enzyme for increasing crop yields and contributes to functional studies of IAA glycoconjugate hydrolases in auxin homeostasis."
Authors: Shannon A. Stirling, Angelica M. Guercio, Ryan M. Patrick, Xing-Qi Huang, Matthew E. Bergman, Varun Dwivedi, Ruy W. J. Kortbeek, Yi-Kai Liu, Fuai Sun, W. Andy Tao, Ying Li, Benoît Boachon, Nitzan Shabek and Natalia Dudareva.
Science (2024)
One-sentence summary: Informational exchange in plants occurs stereospecifically via volatile terpenoids and the KAI2-like signaling pathway.
Editor’s view: Plant responses to chemical signals carried through the air are critical for development, fitness, and adaptation. Stirling et al. identified a mechanism for the perception of the volatile compound (−)-germacrene D through a homolog of the Karrikin receptor KAI2 in petunia pistils. Genetic and biochemical experiments linked this perception with downstream signaling proteins and transcriptional targets. Perturbing the function of some of these components affected volatile-mediated communication and plant fitness.
Abstract: "Plants are constantly exposed to volatile organic compounds (VOCs) that are released during plant-plant communication, within-plant self-signaling, and plant-microbe interactions. Therefore, understanding VOC perception and downstream signaling is vital for unraveling the mechanisms behind information exchange in plants, which remain largely unexplored. Using the hormone-like function of volatile terpenoids in reproductive organ development as a system with a visual marker for communication, we demonstrate that a petunia karrikin-insensitive receptor, PhKAI2ia, stereospecifically perceives the (−)-germacrene D signal, triggering a KAI2-mediated signaling cascade and affecting plant fitness. This study uncovers the role(s) of the intermediate clade of KAI2 receptors, illuminates the involvement of a KAI2ia-dependent signaling pathway in volatile communication, and provides new insights into plant olfaction and the long-standing question about the nature of potential endogenous KAI2 ligand(s)."
Authors: Enyang Mei, Mingliang He, Min Xu, Jiaqi Tang, Jiali Liu, Yingxiang Liu, Zhipeng Hong, Xiufeng Li, Zhenyu Wang, Qingjie Guan, Xiaojie Tian and Qingyun Bu.
Journal of Integrative Plant Biology (2024)
Abstract: "Panicle exsertion is one of the crucial agronomic traits in rice (Oryza sativa). Shortening of panicle exsertion often leads to panicle enclosure and severely reduces seed production. Gibberellin (GA) plays important roles in regulating panicle exsertion. However, the underlying mechanism and the relative regulatory network remain elusive. Here, we characterized the oswrky78 mutant showing severe panicle enclosure, and found that the defect of oswrky78 is caused by decreased bioactive GA contents. Biochemical analysis demonstrates that OsWRKY78 can directly activate GA biosynthesis and indirectly suppress GA metabolism. Moreover, we found OsWRKY78 can interact with and be phosphorylated by mitogen-activated protein kinase (MAPK) kinase OsMAPK6, and this phosphorylation can enhance OsWRKY78 stability and is necessary for its biological function. Taken together, these results not only reveal the critical function of OsWRKY78, but also reveal its mechanism via mediating crosstalk between MAPK and the GA signaling pathway in regulating panicle exsertion."
Authors: Tianshun Zhou, Dong Yu, Liubing Wu, Yusheng Xu, Meijuan Duan and Dingyang Yuan.
Rice Science (2024)
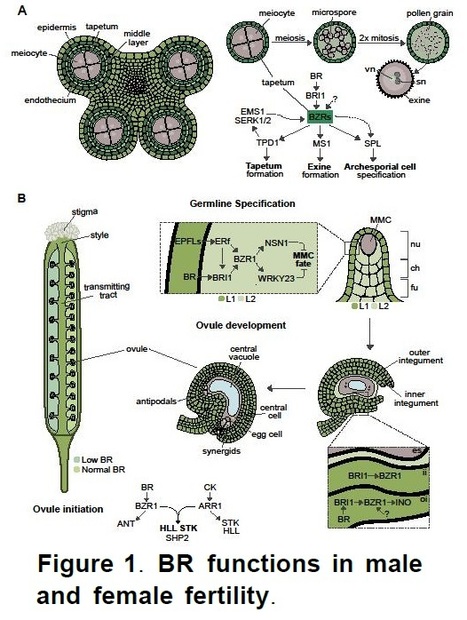
Abstract: "Long-term storage of crop seeds is critical for germplasm resource conservation, food supply and sustainable production. As a major food, rice has a huge stock for production and consumption worldwide, while easily losing food value and seed viability during storage. Thus, understanding the physiological responses and molecular mechanisms of aging tolerance lays the foundation for improving seed storability in rice. This review illustrates the current advances in influential factors, evaluation methods, and identification indexes of seed storability. It also discusses the physiological consequences, molecular mechanisms, and methods to breed aging-tolerant rice in detail. Finally, it points out some challenges in the research of seed storability that need to be addressed in the future. This review provides a theoretical basis and research direction for revealing the mechanisms underlying seed storability and breeding aging tolerant rice."
Authors: Jinyang Lyu, Dongzhi Wang, Na Sun, Fan Yang, Xuepeng Li, Junyi Mu, Runxiang Zhou, Guolan Zheng, Xin Yang, Chenxuan Zhang, Chao Han, Guang-Min Xia, Genying Li, Min Fan, Jun Xiao and Ming-Yi Bai.
Plant Biotechnology Journal (2024)
Abstract: "Regulation of grain size is a crucial strategy for improving the crop yield and is also a fundamental aspect of developmental biology. However, the underlying molecular mechanisms governing grain development in wheat remain largely unknown. In this study, we identified a wheat atypical basic helix–loop–helix (bHLH) transcription factor, TabHLH489, which is tightly associated with grain length through genome-wide association study and map-based cloning. Knockout of TabHLH489 and its homologous genes resulted in increased grain length and weight, whereas the overexpression led to decreased grain length and weight. TaSnRK1α1, the α-catalytic subunit of plant energy sensor SnRK1, interacted with and phosphorylated TabHLH489 to induce its degradation, thereby promoting wheat grain development. Sugar treatment induced TaSnRK1α1 protein accumulation while reducing TabHLH489 protein levels. Moreover, brassinosteroid (BR) promotes grain development by decreasing TabHLH489 expression through the transcription factor BRASSINAZOLE RESISTANT1 (BZR1). Importantly, natural variations in the promoter region of TabHLH489 affect the TaBZR1 binding ability, thereby influencing TabHLH489 expression. Taken together, our findings reveal that the TaSnRK1α1-TabHLH489 regulatory module integrates BR and sugar signalling to regulate grain length, presenting potential targets for enhancing grain size in wheat."
Authors: Xiuying Gao, Jianbo Li, Jing Yin, Yiheng Zhao, Zhongsheng Wu, Lijuan Ma, Baoyi Zhang, Hongsheng Zhang and Ji Huang.
Plant Communications (2024)
Short Summary: qGL3 interacts with OsWDR48 to induce the retention of qGL3 in cytoplasm for degradation, in turn, qGL3 induces the phosphorylation of OsWDR48 at Ser-379 and Ser-386 to weaken their physical interaction. This mechanism helps rice plants quickly respond to BR signals and expands the BR signal transduction network in rice.
Abstract: "Brassinosteroids (BRs) are a class of phytohormones that regulate plant growth and development. In previous studies, we cloned and identified PROTEIN PHOSPHATASE WITH KELCH-LIKE1 (OsPPKL1) as the causal gene for the quantitative trait locus GRAIN LENGHT3 (qGL3) in rice (Oryza sativa). We also showed that qGL3/OsPPKL1 is mainly located in the cytoplasm and nucleus and negatively regulates BR signaling and grain length. Since qGL3 is a negative regulator of BR signaling, its turnover is critical to rapidly respond to changes in BRs. Here, we demonstrate that a WD40 domain-containing protein WD40-REPEAT PROTEIN48 (OsWDR48), which contains a nucleus export signal (NES). The NES signal is crucial for the cytosolic localization of OsWDR48 and also functions in the self-turnover of qGL3. We show that OsWDR48 physically interacts with and genetically acts through qGL3 to modulate BR signaling. Moreover, qGL3 may indirectly promote the phosphorylation of OsWDR48 at Ser-379 and Ser-386 sites. The substitutions of both phosphorylation sites in OsWDR48 to non-phosphorylatable alanine enhanced the strength of the OsWDR48–qGL3 interaction. Furthermore, we found brassinolide could promote the accumulation of non-phosphorylated OsWDR48, leading to strong interaction intensity between qGL3 and OsWDR48. Taken together, OsWDR48 facilitates qGL3 retention and induces degradation of qGL3 in the cytoplasm. These findings suggest that qGL3 self-modulates its turnover by binding to OsWDR48 to regulate its cytoplasmic location and stability, leading to an efficient orchestration of BR signal transduction in rice."
Authors: Mingfeng Zhang, Xiao Luo, Wei He, Min Zhang, Zhirong Peng, Huafeng Deng and Junjie Xing.
Plants (2024)
Abstract: "JAZ proteins function as transcriptional regulators that form a jasmonic acid–isoleucine (JA-Ile) receptor complex with coronatine insensitive 1 (COI1) and regulate plant growth and development. These proteins also act as key mediators in signal transduction pathways that activate the defense-related genes. Herein, the role of OsJAZ4 in rice blast resistance, a severe disease, was examined. The mutation of OsJAZ4 revealed its significance in Magnaporthe oryzae (M. oryzae) resistance and the seed setting rate in rice. In addition, weaker M. oryzae-induced ROS production and expression of the defense genes OsO4g10010, OsWRKY45, OsNAC4, and OsPR3 was observed in osjaz4 compared to Nipponbare (NPB); also, the jasmonic acid (JA) and gibberellin4 (GA4) content was significantly lower in osjaz4 than in NPB. Moreover, osjaz4 exhibited a phenotype featuring a reduced seed setting rate. These observations highlight the involvement of OsJAZ4 in the regulation of JA and GA4 content, playing a positive role in regulating the rice blast resistance and seed setting rate."
Authors: Daniela Barro-Trastoy, Maria Dolores Gomez, Pablo Tornero and Miguel A. Perez-Amador.
Journal of Plant Growth Regulation (2024)
Abstract: "As seed precursors, ovules are fundamental organs during the plant life cycle. Decades of morphological and molecular study have allowed for the elucidation of the complex and intricate genetic network regulating ovule development. Ovule and seed number is highly dependent on the number of ovule primordia that are determined from the placenta during early pistil development. Ovule initiation is positively regulated by the plant hormones auxins, cytokinins, and brassinosteroids, as well as negatively regulated by gibberellins. Each hormone does not act independently; multiple points of hormonal crosstalk occur to coordinately regulate ovule primordia initiation. In this review, we highlight the roles of these hormones and their interactions in the genetic and hormonal network co-regulating ovule initiation in Arabidopsis."
Authors: Jingye Cheng, Yong Jia, Camilla Hill, Tianhua He, Ke Wang, Ganggang Guo, Sergey Shabala, Meixue Zhou, Yong Han and Chengdao Li.
Journal of Advanced Research (2024)
Highlights • Genes concoding gibberellin 2-oxidase are expanded to ten copies in the barley genome and shown copy number variation in the barley pan genome. • The ten genes are transcriptionally expressed in a tissue-specific way, indicating divergent biological functions. • Functions of two genes of gibberellin 2-oxidase were validated by virus-induced gene silencing and CRISPR gene editing to control different agronomic traits. • A natural mutant for the gene was identified with semidwarf stature, early flowering and heavier seeds. • The results provide a promising strategy to minimise the adverse effects of the original Green Revolution semi-dwarf genes to develop the next generation of barley cultivars better adapted to a changing climate.
Abstract: "Introduction - Gibberellin (GA) is a vital phytohormone in regulating plant growth and development. During the “Green Revolution”, modification of GA-related genes created semi-dwarfing phenotype in cereal crops but adversely affected grain weight. Gibberellin 2-oxidases (GA2oxs) in barley act as key catabolic enzymes in deactivating GA, but their functions are still less known. Objectives - This study investigates the physiological function of two HvGA2ox genes in barley and identifies novel semi-dwarf alleles with minimum impacts on other agronomic traits. Methods - Virus-induced gene silencing and CRISPR/Cas9 technology were used to manipulate gene expression of HvGA2ox9 and HvGA2ox8a in barley and RNA-seq was conducted to compare the transcriptome between wild type and mutants. Also, field trials in multiple environments were performed to detect the functional haplotypes. Results - There were ten GA2oxs that distinctly expressed in shoot, tiller, inflorescence, grain, embryo and root. Knockdown of HvGA2ox9 did not affect plant height, while ga2ox8a mutants generated by CRISPR/Cas9 increased plant height and significantly altered seed width and weight due to the increased bioactive GA4 level. RNA-seq analysis revealed that genes involved in starch and sucrose metabolism were significantly decreased in the inflorescence of ga2ox8a mutants. Furthermore, haplotype analysis revealed one naturally occurring HvGA2ox8a haplotype was associated with decreased plant height, early flowering and wider and heavier seed. Conclusion - Our results demonstrate the potential of manipulating GA2ox genes to fine tune GA signalling and biofunctions in desired plant tissues and open a promising avenue for minimising the trade-off effects of Green Revolution semi-dwarfing genes on grain size and weight. The knowledge will promote the development of next generation barley cultivars with better adaptation to a changing climate."
|
Authors: Wenli Hu, Rong Wang, Xiaohua Hao, Shaozhuang Li, Xinjie Zhao, Zijing Xie, Sha Wu, Liqun Huang, Ying Tan, Lianfu Tian and Dongping Li.
The Plant Journal (2024)
Significant Statement: The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight.
Abstract: "A fundamental question in developmental biology is how to regulate grain size to improve crop yields. Despite this, little is still known about the genetics and molecular mechanisms regulating grain size in crops. Here, we provide evidence that a putative protein kinase-like (OsLCD3) interacts with the S-adenosyl-L-methionine synthetase 1 (OsSAMS1) and determines the size and weight of grains. OsLCD3 mutation (lcd3) significantly increased grain size and weight by promoting cell expansion in spikelet hull, whereas its overexpression caused negative effects, suggesting that grain size was negatively regulated by OsLCD3. Importantly, lcd3 and OsSAMS1 overexpression (SAM1OE) led to large and heavy grains, with increased ethylene and decreased polyamines production. Based on genetic analyses, it appears that OsLCD3 and OsSAMS1 control rice grain size in part by ethylene/polyamine homeostasis. The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight."
Authors: Bo Wei and Yuling Jiao.
Seed Biology (2024)
Abstract: "Grain size is a major determinate of bread wheat (Triticum aestivum) yield, which has a broad impact on worldwide food security. Not surprisingly, grain size underwent extensive artificial selection during wheat domestication and breeding. Recent advances in wheat molecular genetics and genomics have facilitated the elucidation of the molecular basis underlying grain size. Grain size determination is the cumulative result of source strength, photoassimilate remobilization, and sink strength. Here, we systematically review the recent progress in the cloning and molecular mechanisms of genes that regulate grain size in wheat following the source to sink flow. In addition, we discuss possible strategies for overcoming the trade-off between grain size and grain number, as well as synergetic improvement of grain yield and grain quality."
Authors: Ray Singh Rathore, Manjari Mishra, Ashwani Pareek and Sneh Lata Singla-Pareek.
Plant Physiology and Biochemistry (2024)
Highlights: • Overexpression of Lonely Guy (OsLOG) gene in rice leads to increase in the active form (trans-zeatin) of cytokinin. • OsLOG mediated elevated cytokinin levels promote ideal plant architecture and rice yield. • Elevated cytokinin levels in OsLOG transgenic lines enhance tolerance to drought and salinity stress and reduce yield penalty through maintenance of redox homeostasis.
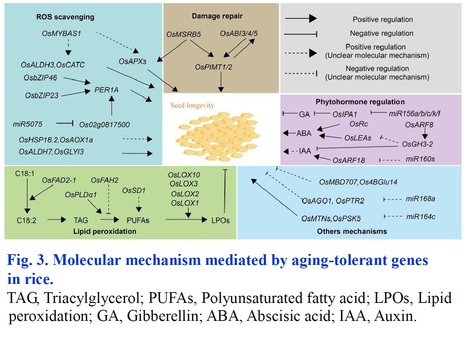
Abstract: "Meristem activity is important for normal plant growth as well as adaptive plastic development under abiotic stresses. Cytokinin has been recognized to have a major role in regulating meristem function which is controlled by cytokinin activating enzymes by fine-tuning the concentrations and spatial distribution of its bioactive forms. It was previously reported that LONELY GUY (LOG) acts in the direct activation pathway of cytokinin in rice shoot meristems. LOG has a cytokinin specific phosphoribohydrolase activity, which transforms inactive cytokinin nucleotides into active free bases. Here, we explored the role of OsLOG in controlling meristem activity mediated by cytokinin and its effects on growth, development, and stress resilience of rice plants. Overexpression of OsLOG in rice led to significant alterations in cytokinin levels in the inflorescence meristem, leading to enhanced plant growth, biomass and grain yield under both non-stress as well as stress conditions such as drought and salinity. Moreover, our study provides insight into how overexpression of OsLOG improves the ability of plants to withstand stress. The OsLOG-overexpressing lines exhibit reduced accumulation of H2O2 along with elevated antioxidant enzyme activities, thereby maintaining better redox homeostasis under stress conditions. This ultimately reduces the negative impact of stresses on grain yield and improves harvest index, as evidenced by observations in the OsLOG-overexpressing lines. In summary, our study emphasizes the diverse role of OsLOG, not only in regulating plant growth and yield via cytokinin but also in enhancing adaptability to abiotic stresses. This highlights its potential to improve crop yield and promote sustainable agriculture."
Authors: Chenyu Rong, Renren Zhang, Yuexin Liu, Zhongyuan Chang, Ziyu Liu, Yanfeng Ding and Chengqiang Ding.
Plant Cell Reports (2024)
Key message: Purine permease PUP11 is essential for rice seed development, regulates the seed setting rate, and influences the cytokinin content, sugar transport, and starch biosynthesis during grain development.
Abstract: "The distribution of cytokinins in plant tissues determines plant growth and development and is regulated by several cytokinin transporters, including purine permease (PUP). Thirteen PUP genes have been identified within the rice genome; however, the functions of most of these genes remain poorly understood. We found that pup11 mutants showed extremely low seed setting rates and a unique filled seed distribution. Moreover, seed formation arrest in these mutants was associated with the disappearance of accumulated starch 10 days after flowering. PUP11 has two major transcripts with different expression patterns and subcellular locations, and further studies revealed that they have redundant positive roles in regulating the seed setting rate. We also found that type-A Response Regulator (RR) genes were upregulated in the developing grains of the pup11 mutant compared with those in the wild type. The results also showed that PUP11 altered the expression of several sucrose transporters and significantly upregulated certain starch biosynthesis genes. In summary, our results indicate that PUP11 influences the rice seed setting rate by regulating sucrose transport and starch accumulation during grain filling. This research provides new insights into the relationship between cytokinins and seed development, which may help improve cereal yield."
Authors: Haihai Wang, Yongcai Huang, Yujie Li, Yahui Cui, Xiaoli Xiang, Yidong Zhu, Qiong Wang, Xiaoqing Wang, Guangjin Ma, Qiao Xiao, Xing Huang, Xiaoyan Gao, Jiechen Wang, Xiaoduo Lu, Brian A. Larkins, Wenqin Wang and Yongrui Wu.
Nature Communications (2024
Editor's view: The mutation of ARFTF17 results in the development of flint kernel architecture in dent maize by reducing excessive pericarp length. This discovery holds significant potential for enhancing grain quality in elite, high-yielding dent maize hybrids.
Abstract: "Dent and flint kernel architectures are important characteristics that affect the physical properties of maize kernels and their grain end uses. The genes controlling these traits are unknown, so it is difficult to combine the advantageous kernel traits of both. We found mutation of ARFTF17 in a dent genetic background reduces IAA content in the seed pericarp, creating a flint-like kernel phenotype. ARFTF17 is highly expressed in the pericarp and encodes a protein that interacts with and inhibits MYB40, a transcription factor with the dual functions of repressing PIN1 expression and transactivating genes for flavonoid biosynthesis. Enhanced flavonoid biosynthesis could reduce the metabolic flux responsible for auxin biosynthesis. The decreased IAA content of the dent pericarp appears to reduce cell division and expansion, creating a shorter, denser kernel. Introgression of the ARFTF17 mutation into dent inbreds and hybrids improved their kernel texture, integrity, and desiccation, without affecting yield."
Source: Agroscope Newsletter (15.02.2024)
Excerpts: "Agroscope has been granted approval by the Federal Office for the Environment for a field trial with spring barley. The focus is on a barley gene that has been disabled by new breeding techniques. The trial, which will be launched in spring 2024 on the Protected Site in Zurich-Reckenholz and will run for three years, aims to determine whether yields can be increased in this manner. The CKX2 gene is involved in the regulation of seed formation. Disabling this gene by means of a new breeding methods (CRISPR/Cas9 genome editing) brings about increased yields in rice and oilseed rape (see, below, ‘From rice to barley’).
"From rice to barley - Crop yield formation is complex and involves many different genes. However, Japanese researchers have discovered that the mutation of the CKX2 gene in rice has an unexpectedly significant effect on yield. Results were so convincing that they are now used in rice breeding. Research results show that genes corresponding to the CKX2 gene from rice also play a role e.g. in oilseed-rape yield formation. Therefore, it is reasonable to study this effect in further crops. In the best-case scenario, at the end of these trials on the Protected Site it will be possible to issue a recommendation as to whether breeders should disable one or both CKX2 genes in order to boost yields. What is certain, however, is that important information will be provided on the function of the CKX2 genes in barley – and hence further pieces of the puzzle will be available for a better understanding of yield formation."
Authors: Qin Zheng, Zhenning Teng, Jianhua Zhang and Nenghui Ye.
Plants (2024)
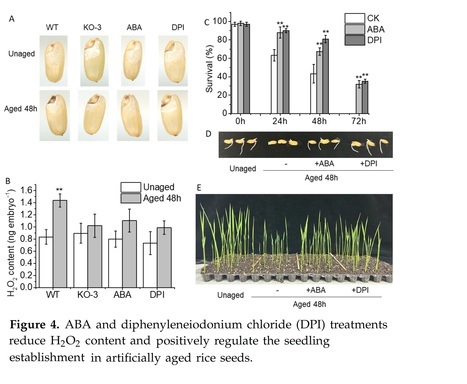
Abstract: "The seed, a critical organ in higher plants, serves as a primary determinant of agricultural productivity, with its quality directly influencing crop yield. Improper storage conditions can diminish seed vigor, adversely affecting seed germination and seedling establishment. Therefore, understanding the seed-aging process and exploring strategies to enhance seed-aging resistance are paramount. In this study, we observed that seed aging during storage leads to a decline in seed vigor and can coincide with the accumulation of hydrogen peroxide (H2O2) in the radicle, resulting in compromised or uneven germination and asynchronous seedling emergence. We identified the abscisic acid (ABA) catabolism gene, abscisic acid 8′-hydroxylase 2 (OsABA8ox2), as significantly induced by aging treatment. Interestingly, transgenic seeds overexpressing OsABA8ox2 exhibited reduced seed vigor, while gene knockout enhanced seed vigor, suggesting its role as a negative regulator. Similarly, seeds pretreated with ABA or diphenyleneiodonium chloride (DPI, an H2O2 inhibitor) showed increased resistance to aging, with more robust early seedling establishment. Both OsABA8ox2 mutant seeds and seeds pretreated with ABA or DPI displayed lower H2O2 content during aging treatment. Overall, our findings indicate that ABA mitigates rice seed aging by reducing H2O2 accumulation in the radicle. This study offers valuable germplasm resources and presents a novel approach to enhancing seed resistance against aging."
Authors: Xiaoxing Zhang, Wenjing Meng, Dapu Liu, Dezhuo Pan, Yanzhao Yang, Zhuo Chen, Xiaoding Ma, Wenchao Yin, Mei Niu, Nana Dong, Jihong Liu, Weifeng Shen, Yuqin Liu, Zefu Lu, Chengcai Chu, Qian Qian, Mingfu Zhao and Hongning Tong.
Science (2024)
One-sentence summary: Cell-specific activation of a brassinosteroid-degrading enzyme results in clustered-spikelet rice and improves grain yield.
Editor's view: Hormones are small mobile signaling molecules that control growth and development. Zhang et al. studied how a hormone signaling pathway acts in a precise spatiotemporal manner (i.e., cell- and tissue-specific signaling) to enhance grain number in rice. They identified a brassinosteroid catabolism gene that is the causative gene underlying enhanced grain number in Clustered-Spikelet rice through a signaling cascade and downstream gene expression of a key transcription factor. This work paves the way to understanding how spatial regulation of brassinosteroid hormonal pathways in secondary branch meristems enhances panicle branching and grain number, crucial traits for enhancing rice yields.
Abstract: "Crop yield potential is constrained by the inherent trade-offs among traits such as between grain size and number. Brassinosteroids (BRs) promote grain size, yet their role in regulating grain number is unclear. By deciphering the clustered-spikelet rice germplasm, we show that activation of the BR catabolic gene BRASSINOSTEROID-DEFICIENT DWARF3 (BRD3) markedly increases grain number. We establish a molecular pathway in which the BR signaling inhibitor GSK3/SHAGGY-LIKE KINASE2 phosphorylates and stabilizes OsMADS1 transcriptional factor, which targets TERMINAL FLOWER1-like gene RICE CENTRORADIALIS2. The tissue-specific activation of BRD3 in the secondary branch meristems enhances panicle branching, minimizing negative effects on grain size, and improves grain yield. Our study showcases the power of tissue-specific hormonal manipulation in dismantling the trade-offs among various traits and thus unleashing crop yield potential in rice."
Authors: Júlia de Paiva Gonçalves, Karla Gasparini, Edgard T. Picoli, Maximiller Dal-Bianco Costa, Wagner L. Araujo, Agustin Zsögön and Dimas M. Ribeiro.
Journal of Plant Physiology (2024)
Abstract: "Seed development, dormancy, and germination are connected with changes in metabolite levels. Not surprisingly, a complex regulatory network modulates biosynthesis and accumulation of storage products. Seed development has been studied profusely in Arabidopsis thaliana and has provided valuable insights into the genetic control of embryo development. However, not every inference applies to crop legumes, as these have been domesticated and selected for high seed yield and specific metabolic profiles and fluxes. Given its enormous economic relevance, considerable work has contributed to shed light on the mechanisms that control legume seed growth and germination. Here, we summarize recent progress in the understanding of regulatory networks that coordinate seed metabolism and development in legumes."
Authors: Xiaofeng Luo, Yujia Dai, Baoshan Xian, Jiahui Xu, Ranran Zhang, Muhammad Saad Rehmani, Chuan Zheng, Xiaoting Zhao, Kaitao Mao, Xiaotong Ren, Shaowei Wei, Lei Wang, Juan He, Weiming Tan, Junbo Du, Weiguo Liu, Shu Yuan and Kai Shu.
Journal of Integrative Plant Biology (2024)
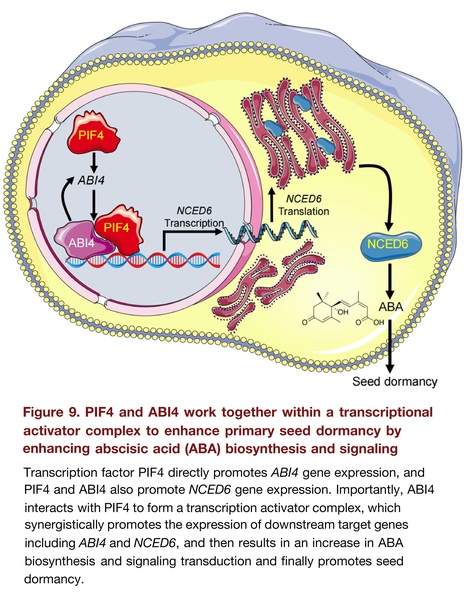
Abstract: "Transcriptional regulation plays a key role in the control of seed dormancy, and many transcription factors (TFs) have been documented. However, the mechanisms underlying the interactions between different TFs within a transcriptional complex regulating seed dormancy remain largely unknown. Here, we showed that TF PHYTOCHROME-INTERACTING FACTOR4 (PIF4) physically interacted with the abscisic acid (ABA) signaling responsive TF ABSCISIC ACID INSENSITIVE4 (ABI4) to act as a transcriptional complex to promote ABA biosynthesis and signaling, finally deepening primary seed dormancy. Both pif4 and abi4 single mutants exhibited a decreased primary seed dormancy phenotype, with a synergistic effect in the pif4/abi4 double mutant. PIF4 binds to ABI4 to form a heterodimer, and ABI4 stabilizes PIF4 at the protein level, whereas PIF4 does not affect the protein stabilization of ABI4. Subsequently, both TFs independently and synergistically promoted the expression of ABI4 and NCED6, a key gene for ABA anabolism. The genetic evidence is also consistent with the phenotypic, physiological and biochemical analysis results. Altogether, this study revealed a transcriptional regulatory cascade in which the PIF4–ABI4 transcriptional activator complex synergistically enhanced seed dormancy by facilitating ABA biosynthesis and signaling."
Authors: Masahiko Otani, Ryo Tojo, Sarah Regnard, Lipeng Zheng, Takumi Hoshi, Suzuha Ohmori, Natsuki Tachibana, Tomohiro Sano, Shizuka Koshimizu, Kazuya Ichimura, Jean Colcombet and Naoto Kawakami.
bioRxiv (2024)
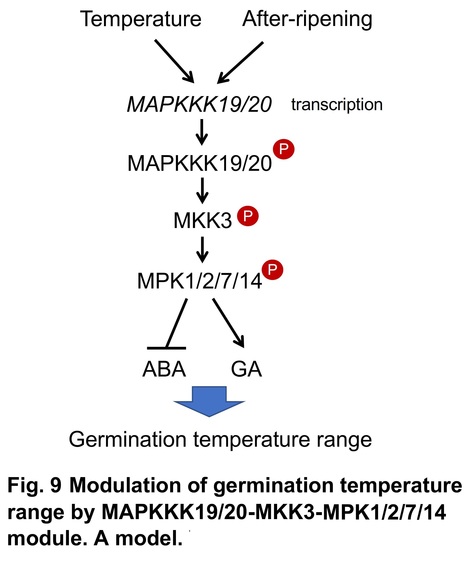
Abstract: "Temperature is a major environmental cue for seed germination. The permissive temperature range for germination is narrow in dormant seeds and expands during after-ripening. Quantitative trait loci analyses of pre-harvest sprouting in cereals have revealed that MKK3, a mitogen-activated protein kinase (MAPK) cascade protein, is a negative regulator of grain dormancy. Here we show that the MAPKKK19/20-MKK3-MPK1/2/7/14 cascade modulates germination temperature range in Arabidopsis seeds by elevating germinability of the seeds at sub- and supra-optimal temperatures. The expression of MAPKKK19 and MAPKKK20 is regulated by an unidentified temperature sensing and signaling mechanism the sensitivity of which is modulated during after-ripening of the seeds, and MPK7 is activated at the permissive temperature for germination regulated by expression levels of MAPKKK19/20. Activation of the MKK3 cascade represses abscisic acid (ABA) biosynthesis enzyme gene expression, and induces expression of ABA catabolic enzyme and gibberellic acid biosynthesis enzyme genes, resulting in expansion of the germinable temperature range. Our data demonstrate that the MKK3 cascade integrates temperature and after-ripening signals to germination processes including phytohormone metabolism."
Authors: Daxia Wu, Yanan Cao, Daojian Wang, Guoxinan Zong, Kunxu Han, Wei Zhang, Yanhua Qi, Guohua Xu and Yali Zhang.
Plant Physiology (2024)
One-sentence summary: Auxin transport inhibitor response 1 (TIR1) plays an essential role in rice grain yield and quality via modulating sugar transport into the endosperm.
Abstract: "Cereal endosperm represents the most important source of the world's food. Nevertheless, the molecular mechanisms behind sugar import into rice (Oryza sativa) endosperm and their relationship with auxin signaling are poorly understood. Here, we report that auxin transport inhibitor response 1 (TIR1) plays an essential role in rice grain yield and quality via modulating sugar transport into endosperm. The fluctuations of OsTIR1 transcripts parallel to the early stage of grain expansion among those of five TIR1/AFB (AFB, auxin-signaling F-Box) auxin co-receptor proteins. OsTIR1 is abundantly expressed in ovular vascular trace, nucellar projection, nucellar epidermis, aleurone layer cells, and endosperm, providing a potential path for sugar into the endosperm. Compared to wild-type (WT) plants, starch accumulation is repressed by mutation of OsTIR1 and improved by overexpression of the gene, ultimately leading to reduced grain yield and quality in tir1 mutants but improvement in overexpression lines. Of rice AUXIN RESPONSE FACTOR (ARF) genes, only the OsARF25 transcript is repressed in tir1 mutants and enhanced by overexpression of OsTIR1; its highest transcript is recorded at 10 d after fertilization, consistent with OsTIR1 expression. Also, OsARF25 can bind the promoter of sugar transporter OsSWEET11 (SWEET11, sugars will eventually be exported transporter) in vivo and in vitro. arf25 and arf25/sweet11 mutants exhibit reduced starch content and seed size (relative to the WTs), similar to tir1 mutants. Our data reveal that OsTIR1 mediates sugars import into endosperm via the auxin signaling component OsARF25 interacting with sugar transporter OsSWEET11. The results of this study are of great significance to further clarify the regulatory mechanism of auxin signaling on grain development in rice."
|

 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...