Your new post is loading...
 Your new post is loading...
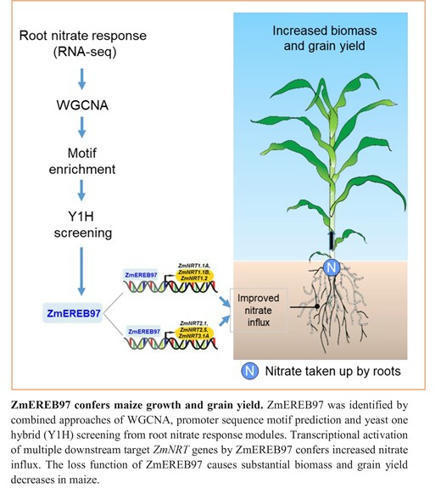
Authors: s Qi Wu, Jinyan Xu, Yingdi Zhao, Yuancong Wang, Ling Zhou, Lihua Ning, Sergey Shabala and Han Zhao.
Plant Physiology (2024)
One-sentence summary: An ethylene response transcription factor plays an important role in the nitrogen-signaling network and in optimizing nitrate uptake in maize.
Abstract: "Maize (Zea mays L.) has very strong requirements for nitrogen. However, the molecular mechanisms underlying the regulations of nitrogen uptake and translocation in this species are not fully understood. Here, we report that an APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor ZmEREB97 functions as an important regulator in the N-signaling network in maize. Predominantly expressed and accumulated in main root and lateral root primordia, ZmEREB97 rapidly responded to nitrate treatment. By overlapping the analyses of differentially expressed genes and conducting a DAP-seq assay, we identified 1446 potential target genes of ZmEREB97. Among these, 764 genes were co-regulated in two lines of zmereb97 mutants. Loss of function of ZmEREB97 substantially weakened plant growth under both hydroponic and soil conditions. Physiological characterization of zmereb97 mutant plants demonstrated that reduced biomass and grain yield were both associated with reduced nitrate influx, decreased nitrate content and less N accumulation. We further demonstrated that ZmEREB97 directly targets and regulates the expression of six ZmNRT genes by binding to the GCC box-related sequences in gene promoters. Collectively, these data suggest that ZmEREB97 is a major positive regulator of the nitrate response and that it plays an important role in optimizing nitrate uptake, offering a target for improvement of nitrogen use efficiency in crops."
Authors: Jingyi Han (撖静宜), Thomas Welch, Ute Voß, Teva Vernoux, Rahul Bhosale and Anthony Bishopp.
iScience (2024)
Highlights • The first intron of ARF7 is required for expression in the root apical meristem. • It is not required for expression in either lateral roots or the shoot apex. • We find no binding sites in the first intron required for expression in root tips. • We propose that this intron exerts its effects via Intron Mediated Enhancement.
Abstract: "Auxin regulates plant growth and development through the transcription factors of the AUXIN RESPONSE FACTOR (ARF) gene family. ARF7 is one of five activators that bind DNA and elicit downstream transcriptional responses. In roots, ARF7 regulates growth, gravitropism and redundantly with ARF19, lateral root organogenesis. In this study we analyzed ARF7 cis-regulation, using different non-coding sequences of the ARF7 locus to drive GFP. We show that constructs containing the first intron led to increased signal in the root tip. Although bioinformatics analyses predicted several transcription factor binding sites in the first intron, we were unable to significantly alter expression of GFP in the root by mutating these. We instead observed the intronic sequences needed to be present within the transcribed sequences to drive expression in the root meristem. These data support a mechanism by which Intron Mediated Enhancement regulates the tissue specific expression of ARF7 in the root meristem."
Authors: Feimei Guo, Minghui Lv, Jingjie Zhang and Jia Li.
Plant and Cell Physiology (2024)
Abstract: "Brassinosteroids (BRs) are a group of polyhydroxylated phytosterols that play essential roles in regulating plant growth and development as well as stress adaptation. It is worth noting that BRs do not function alone, but rather they crosstalk with other endogenous signaling molecules, including the phytohormones auxin, cytokinins (CKs), gibberellins (GAs), abscisic acid (ABA), ethylene (ET), jasmonates (JAs), salicylic acid (SA), and strigolactones (SLs), forming elaborate signaling networks to modulate plant growth and development. BRs interact with other phytohormones mainly by regulating each others’ homeostasis, transport, or signaling pathway at the transcriptional and posttranslational levels. In this review, we focus our attention on current research progress in BR signal transduction and the crosstalk between BRs and other phytohormones."
Authors: Da Zhang, Tan He, Xumin Wang, Chenchen Zhou, Youpeng Chen, Xin Wang, Shixiang Wang, Shuangcheng He, Yuan Guo, Zijin Liu and Mingxun Chen.
Plant Physiology (2024)
One-sentence summary: A MYB-like transcription factor positively modulates seed germination in response to salinity stress by regulating the expression of germination-associated genes.
Abstract: "Seed germination is a critical checkpoint for plant growth under unfavorable environmental conditions. In Arabidopsis (Arabidopsis thaliana), the abscisic acid (ABA) and gibberellic acid (GA) signaling pathways play important roles in modulating seed germination. However, the molecular links between salinity stress and ABA/GA signaling are not well understood. Herein, we showed that the expression of DIVARICATA1 (DIV1), which encodes a MYB-like transcription factor, was induced by GA and repressed by ABA, salinity, and osmotic stress in germinating seeds. DIV1 positively regulated seed germination in response to salinity stress by directly regulating the expression of DELAY OF GERMINATION 1-LIKE 3 (DOGL3) and GA-STIMULATED ARABIDOPSIS 4 (GASA4) and indirectly regulating the expression of several germination-associated genes. Moreover, NUCLEAR FACTOR-YC9 (NF-YC9) directly repressed the expression of DIV1 in germinating seeds in response to salinity stress. These results help reveal the function of the NF-YC9–DIV1 module and provide insights into the regulation of ABA and GA signaling in response to salinity stress during seed germination in Arabidopsis."
Authors: Sonia Gazzarrini and Liang Song.
Annual Review of Plant Biology (2024)
Abstract: "Development is a chain reaction in which one event leads to another until the completion of a life cycle. Phase transitions are milestone events in the cycle of life. LEAFY COTYLEDON1 (LEC1), ABA INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 proteins, collectively known as LAFL, are master transcription factors (TFs) regulating seed and other developmental processes. Since the initial characterization of the LAFL genes (44, 71, 98–100), more than three decades of active research has generated tremendous amounts of knowledge about these TFs, whose roles in seed development and germination have been comprehensively reviewed (63, 43, 65, 81). Recent advances in cell biology and genetic and genomic tools have allowed the characterization of the LAFL regulatory networks in previously challenging tissues at a higher throughput and resolution in reference species and crops. In this review, we provide a holistic perspective by integrating advances at the epigenetic, transcriptional, posttranscriptional, and protein levels to exemplify the spatiotemporal regulation of the LAFL networks in Arabidopsis seed development and phase transitions, and we briefly discuss the evolution of these TF networks."
Authors: Amanpreet Kaur, Norman B. Best, Thomas Hartwig, Josh Budka, Rajdeep Khangura, Steven McKenzie, Alejandro Aragón-Raygoza, Josh Strable, Burkhard Schulz and Brian P. Dilkes.
Plant Physiology (2023)
One-sentence summary: Molecular identity of a maize semidwarf mutant reveals a role for the maize GRAS domain transcription factor ortholog of DWARF AND LOW TILLERING in brassinosteroid signaling.
Abstract: "Brassinosteroids (BR) and gibberellins (GA) regulate plant height and leaf angle in maize (Zea mays). Mutants with defects in BR or GA biosynthesis or signaling identify components of these pathways and enhance our knowledge about plant growth and development. In this study, we characterized three recessive mutant alleles of GRAS transcription factor 42 (gras42) in maize, a GRAS transcription factor gene orthologous to the DWARF AND LOW TILLERING (DLT) gene of rice (Oryza sativa). These maize mutants exhibited semi-dwarf stature, shorter and wider leaves, and more upright leaf angle. Transcriptome analysis revealed a role for GRAS42 as a determinant of BR signaling. Analysis of the expression consequences from loss of GRAS42 in the gras42-mu1021149 mutant indicated a weak loss of BR signaling in the mutant, consistent with its previously demonstrated role in BR signaling in rice. Loss of BR signaling was also evident by the enhancement of weak BR biosynthetic mutant alleles in double mutants of nana plant1-1 and gras42-mu1021149. The gras42-mu1021149 mutant had little effect on GA-regulated gene expression, suggesting that GRAS42 is not a regulator of core GA signaling genes in maize. Single cell expression data identified gras42 expressed among cells in the G2/M phase of the cell cycle consistent with its previously demonstrated role in cell cycle gene expression in Arabidopsis (Arabidopsis thaliana). Cis-acting natural variation controlling GRAS42 transcript accumulation was identified by expression genome-wide association study (eGWAS) in maize. Our results demonstrate a conserved role for GRAS42/SCARECROW-LIKE 28 (SCL28)/DLT in BR signaling, clarify the role of this gene in GA signaling, and suggest mechanisms of tillering and leaf angle control by BR."
Authors: Tomás Urzúa Lehuedé, Victoria Berdion Gabarain, Tomas Moyano, Lucia Ferrero, Miguel Angel Ibeas, Hernan Salinas-Grenet, Gerardo Núñez-Lillo, Romina Acha, Jorge Perez, Florencia Perotti, Virginia Natali Miguel, Fiorella Paola Spies, Miguel A. Rosas, Michitaro Shibata, Diana R. Rodríguez-García, Adrian A. Moreno, Keiko Sugimoto, Karen Sanguinet, Claudio Meneses, Raquel L. Chan, Federico Ariel, Jose M. Alvarez and José M. Estevez.
bioRxiv (2024)
Abstract: "The root hair (RH) cells can elongate to several hundred times their initial size, and are an ideal model system for investigating cell size control. Their development is influenced by both endogenous and external signals, which are combined to form a integrative response. Surprisingly, a low temperature condition of 10°C causes an increased RH growth in Arabidopsis and in several monocots, even when the development of the rest of the root and aerial parts of the plant are halted. Previously, we demonstrated a strong correlation between the growth response and a significant decrease in nutrient availability in the medium under low temperature conditions. However, the molecular basis responsible for receiving and transmitting signals related to the availability of nutrients in the soil, and their relation to plant development, remain largely unknown. We decided to further investigate the intricate molecular processes behind the particular responsiveness of this root cell type at low temperature. In this study, we have discovered a gene regulatory network (GRN) controlling early transcriptome responses to low temperature. This GNR is commanded by specific transcription factors (FTs), namely ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), a member of the homeodomain leucine zipper (HD-Zip I) group I 13 (AtHB13), the trihelix TF GT2-LIKE1 (GTL1), and a previously unidentified MYB-like TF (AT2G01060). Furthermore, we have identified four downstream RSL4 targets AtHB16, AtHB23, EARLY-RESPONSIVE TO DEHYDRATION 7 (ERD7) and ERD10 suggesting their participation in the regulation of RH development under these conditions. Functional analysis shows that such components of the RSL4-dependent transcriptional cascade influence the subsequent RH growth response to low temperature. These discoveries enhance our comprehension of how plants synchronize the RH growth in response to variations in temperature and nutrient availability at the cellular level."
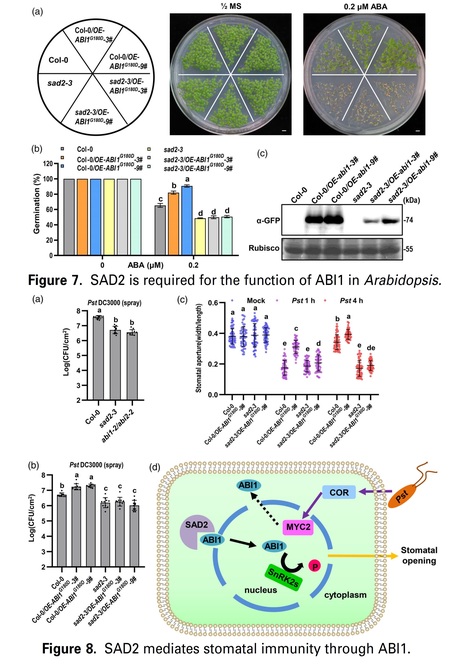
Authors: Lu Liu, Yanzhi Liu, Xuehan Ji, Xia Zhao, Jun Liu and Ning Xu.
The Plant Journal (2024)
Significance Statement: Bacterial pathogens secret coronatine (COR) to recruit an importin β protein SAD2 for interfering ABA signaling in stomata and promote invasion.
Abstract: "Stomatal immunity plays an important role during bacterial pathogen invasion. Abscisic acid (ABA) induces plants to close their stomata and halt pathogen invasion, but many bacterial pathogens secrete phytotoxin coronatine (COR) to antagonize ABA signaling and reopen the stomata to promote infection at early stage of invasion. However, the underlining mechanism is not clear. SAD2 is an importin β family protein, and the sad2 mutant shows hypersensitivity to ABA. We discovered ABI1, which negatively regulated ABA signaling and reduced plant sensitivity to ABA, was accumulated in the plant nucleus after COR treatment. This event required SAD2 to import ABI1 to the plant nucleus. Abolition of SAD2 undermined ABI1 accumulation. Our study answers the long-standing question of how bacterial COR antagonizes ABA signaling and reopens plant stomata during pathogen invasion."
Authors: Yoon Jeong Jang, Taehoon Kim, Makou Lin, Jeongim Kim, Kevin Begcy, Zhongchi Liu and Seonghee Lee.
bioRxiv (2024)
Abstract: "The plant hormone auxin plays a crucial role in regulating important functions in strawberry fruit development. Although a few studies have described the complex auxin biosynthetic and signaling pathway in wild diploid strawberry (Fragaria vesca), the molecular mechanisms underlying auxin biosynthesis and crosstalk in octoploid strawberry fruit development are not fully characterized. To address this knowledge gap, comprehensive transcriptomic analyses were conducted at different stages of fruit development and compared between the achene and receptacle to identify developmentally regulated auxin biosynthetic genes and transcription factors during the fruit ripening process. Similar to wild diploid strawberry, octoploid strawberry accumulates high levels of auxin in achene compared to receptacle. Consistently, genes functioning in auxin biosynthesis and conjugation, such as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAAs), YUCCA (YUCs), and GRETCHEN HAGEN 3 (GH3s) were found to be primarily expressed in the achene, with low expression in the receptacle. Interestingly, several genes involved in auxin transport and signaling like PIN-FORMED (PINs), AUXIN/INDOLE-3-ACETIC ACID proteins (Aux/IAAs), TRANSPORT INHIBITOR RESPONSE 1 / AUXIN-SIGNALING F-BOX (TIR/AFBs) and AUXIN RESPONSE FACTOR (ARFs) were more abundantly expressed in the receptacle. Moreover, by examining DEGs and their transcriptional profiles across all six developmental stages, we identified key auxin-related genes co-clustered with transcription factors from the NAM-ATAF1,2-CUC2/ WRKYGQK motif (NAC/WYKY), BASIC REGION/ LEUCINE ZIPPER motif (bZIP), and APETALA2/Ethylene Responsive Factor (AP2/ERF) groups. These results elucidate the complex regulatory network of auxin biosynthesis and its intricate crosstalk within the achene and receptacle, enriching our understanding of fruit development in octoploid strawberries."
Authors: Yuming Sun and Alisdair R. Fernie.
Trends in Plant Science (2024)
Highlights Plant secondary metabolism plays a paramount role in plant adaptation to global climate change, a comprehensive understanding of the interplay between plant secondary metabolism and climate variations favors crop improvement in a climate-fluctuating world. Climate changes exert diverse effects on plant secondary metabolism, determined by factors such as metabolite type, stress intensity, and plant species. Combinations of two individual climatic factors can result in an amplifying, neutralizing, or one-factor-dominated consequence, but our knowledge of these consequences is limited. Resource allocation and growth-defense trade-off hypothesis partially explain how plants respond to climate changes through secondary metabolism at the ecological level, while hormone signaling, hormones, and transcriptional and post-transcriptional regulation networks are involved with the molecular mechanisms underlying these responses.
Abstract: "Climate changes have unpredictable effects on ecosystems and agriculture. Plants adapt metabolically to overcome these challenges, with plant secondary metabolites (PSMs) being crucial for plant–environment interactions. Thus, understanding how PSMs respond to climate change is vital for future cultivation and breeding strategies. Here, we review PSM responses to climate changes such as elevated carbon dioxide, ozone, nitrogen deposition, heat and drought, as well as a combinations of different factors. These responses are complex, depending on stress dosage and duration, and metabolite classes. We finally identify mechanisms by which climate change affects PSM production ecologically and molecularly. While these observations provide insights into PSM responses to climate changes and the underlying regulatory mechanisms, considerable further research is required for a comprehensive understanding."
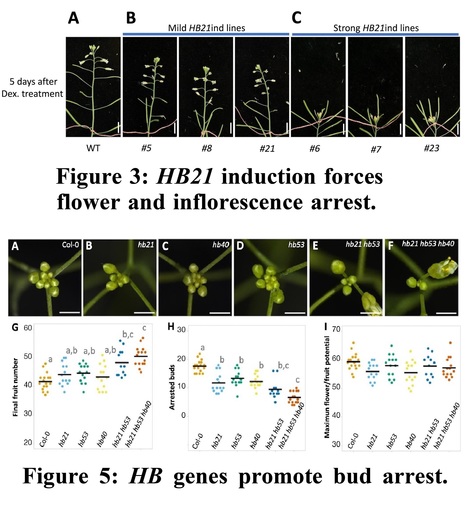
Authors: Verónica Sánchez-Gerschon, Irene Martínez-Fernández, María R. González-Bermúdez, Sergio de la Hoz-Rodríguez, Florenci V. González, Jorge Lozano-Juste, Cristina Ferrándiz and Vicente Balanzà.
Plant Physiology (2024)
One-sentence summary: Three homeodomain leucine zipper transcription factors trigger abscisic acid accumulation in the inflorescence apex at the end of the reproductive phase, arresting unpollinated floral bud development.
Abstract: "Flowers, and hence, fruits and seeds, are produced by the activity of the inflorescence meristem after the floral transition. In plants with indeterminate inflorescences the final number of flowers produced by the inflorescence meristem is determined by the length of the flowering period, which ends with inflorescence arrest. Inflorescence arrest depends on many different factors, such as the presence of seeds, the influence of the environment, or endogenous factors such as phytohormone levels and age, which modulate inflorescence meristem activity. The FRUITFULL-APETALA2 (FUL-AP2) pathway plays a major role in regulating the end of flowering, likely integrating both endogenous cues and those related to seed formation. Among AP2 targets, HOMEOBOX PROTEIN21 (HB21) has been identified as a putative mediator of AP2 function in the control of inflorescence arrest. HB21 is a homeodomain leucine zipper transcription factor involved in establishing axillary bud dormancy. Here we characterized the role of HB21 in the control of the inflorescence arrest at the end of flowering in Arabidopsis (Arabidopsis thaliana). HB21, together with HB40 and HB53, are upregulated in the inflorescence apex at the end of flowering, promoting floral bud arrest. We also show that abscisic acid (ABA) accumulation occurs in the inflorescence apex in an HB-dependent manner. Our work suggests a physiological role of ABA in floral bud arrest at the end of flowering, pointing to ABA as a regulator of inflorescence arrest downstream of the HB21/40/53 genes."
Author: Leiyun Yang
The Plant Cell (2024)
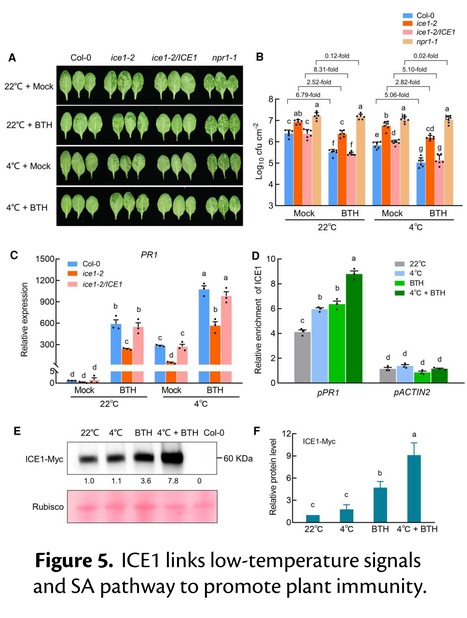
Excerpts: "Simultaneously, low temperature stimulates plant immunity by activating salicylic acid (SA)-mediated signaling (Wu et al. 2019). However, the mechanism by which plants integrate cold signals and immune signaling remain elusive. To investigate this question, Shaoqin Li and colleagues (Li et al. 2024) studied the potential involvement of ICE1 in cold-induced immunity in Arabidopsis."
"To elucidate how ICE1 regulates cold-induced immunity, the authors conducted a yeast-two-hybrid screen and identified NON-EXPRESSER OF PR GENES 1 (NPR1), a master transcriptional co-activator of PATHOGENESIS-RELATED GENE 1 (PR1) in SA-activated immunity, as an ICE1 interactor.....Additionally, the authors found that ICE1 directly binds to the PR1 promoter for gene activation. These results illustrate that ICE1 interacts with NPR1 and is required for SA-mediated immunity by directly promoting PR1 expression."
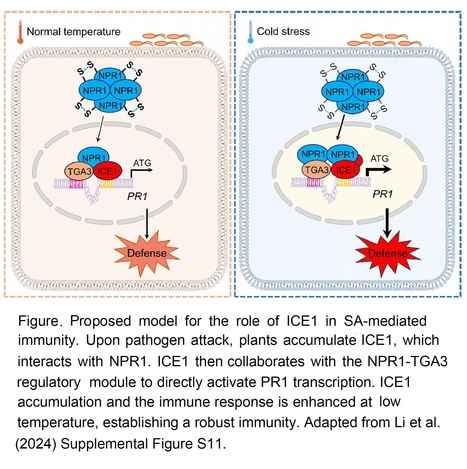
"These results support the conclusion that ICE1 and TGA3 work synergistically to promote PR1 transcription. This study not only shed light on a new role of ICE1 in SA-mediated immunity at low temperature, but also revealed NPR1-TGA3/ICE1 as an important nexus integrating SA signaling and cold signals in plant immunity (see Figure)."
Authors: Tao Zhang, Xinyu Wang, Yanchao Yuan, Shoujie Zhu, Chunying Liu, Yuxi Zhang, Shupeng Gai.
Horticulture Research (2024)
Abstract: "Bud endodormancy in perennial plants is a sophisticated system that adapts to seasonal climatic changes. Growth-promoting signals such as low temperature and gibberellins (GAs) are crucial for facilitating budbreak following endodormancy release (EDR). However, the regulatory mechanisms underlying GA-mediated budbreak in tree peony (Paeonia suffruticosa) remain unclear. In tree peony, the expression of PsmiR159b among three differentially expressed miR159 members was inhibited with the prolonged chilling, and overexpression of PsMIR159b delayed budbreak, whereas silencing PsmiR159b promoted budbreak after dormancy. PsMYB65, a downstream transcription factor in the GA pathway, was induced by prolonged chilling and exogenous GA3 treatments. PsMYB65 was identified as a target of PsmiR159b, and promoted budbreak in tree peony. RNA-seq of PsMYB65-silenced buds revealed significant enrichment in the GO terms regulation of ‘cell cycle’ and ‘DNA replication’ among differentially expressed genes. Yeast one-hybrid and electrophoretic mobility shift assays demonstrated that PsMYB65 directly bound to the promoter of the type-D cyclin gene PsCYCD3;1. Dual-luciferase reporter assay indicated that PsMYB65 positively regulate PsCYCD3;1 expression, suggesting that miR159b-PsMYB65 module contributes to budbreak by influencing the cell cycle. Our findings revealed that the PsmiR159b-PsMYB65 module functioned in budbreak after dormancy by regulating cell proliferation, providing valuable insights into the endodormancy release regulation mechanism."
|
Authors: Zhaoqing Song, Yeting Bian, Yuntao Xiao and Dongqing Xu.
Journal of Plant Physiology (2024)
Abstract: "B-box containing proteins (BBXs) are a class of zinc-ligating transcription factors or regulators that play essential roles in various physiological and developmental processes in plants. They not only directly associate with target genes to regulate their transcription, but also interact with other transcription factors to mediate target genes’ expression, thus forming a complex transcriptional network ensuring plants’ adaptation to dynamically changing light environments. This review summarizes and highlights the molecular and biochemical properties of BBXs, as well as recent advances with a focus on their critical regulatory functions in photomorphogenesis (de-etiolation), shade avoidance, photoperiodic-mediated flowering, and secondary metabolite biosynthesis and accumulation in plants."
Authors: Byron Rusnak, Frances K. Clark, Batthula Vijaya Lakshmi Vadde and Adrienne H.K. Roeder.
Annual Review of Cell and Developmental Biology (2024)
Abstract: "One of the fundamental questions in developmental biology is how a cell is specified to differentiate as a specialized cell type. Traditionally, plant cell types were defined based on their function, location, morphology, and lineage. Currently, in the age of single-cell biology, researchers typically attempt to assign plant cells to cell types by clustering them based on their transcriptomes. However, because cells are dynamic entities that progress through the cell cycle and respond to signals, the transcriptome also reflects the state of the cell at a particular moment in time, raising questions about how to define a cell type. We suggest that these complexities and dynamics of cell states are of interest and further consider the roles signaling, stochasticity, cell cycle, and mechanical forces play in plant cell fate specification. Once established, cell identity must also be maintained. With the wealth of single-cell data coming out, the field is poised to elucidate both the complexity and dynamics of cell states."
Authors: Baolei Li and Jiaqi Sun.
Plant Physiology (2024)
Excerpt: "The functions of DIVARICATA (DIV) proteins, which belong to the MYB-like TF family, remain elusive (Fang et al., 2018). Six DIV genes have been identified in Arabidopsis based on their structural resemblance to the tomato I-box binding factor SlMYBI (Machemer et al., 2011). Among these, only DIV2 has been characterized as a positive regulator of seed germination in response to ABA and salt stress (Fang et al., 2018). However, the roles of the other DIV genes in the process of seed germination in response to salinity stress remain largely unexplored. In this issue of Plant Physiology, Zhang et al. (2024) provide evidence that DIV1 functions as a crucial integrative regulator that links salinity stress with ABA/GA signaling during seed germination."
"In summary, Zhang et al. (2024) illustrate the significant role of DIV1 as a positive regulator of Arabidopsis seed germination in response to salinity stress. DIV1 exerts a direct influence on ABA/GA signaling by promoting GASA4 expression and suppressing DOGL3 expression. Additionally, it indirectly modulates the expression of a series of germination-associated genes.
Authors: Davide Marzi, Patrizia Brunetti, Shashank Sagar Saini, Gitanjali Yadav, Giuseppe Diego Puglia and Raffaele Dello Ioio.
Frontiers in Genetics (2024)
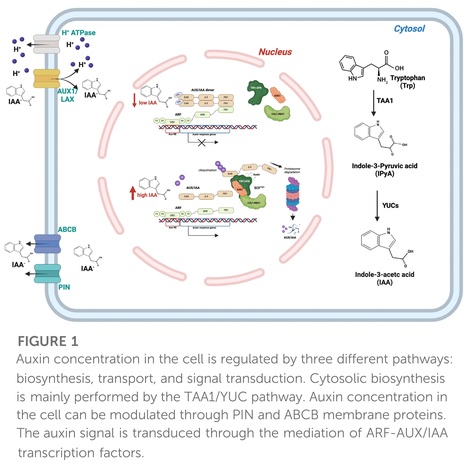
Abstract: "Global climate change (GCC) is posing a serious threat to organisms, particularly plants, which are sessile. Drought, salinity, and the accumulation of heavy metals alter soil composition and have detrimental effects on crops and wild plants. The hormone auxin plays a pivotal role in the response to stress conditions through the fine regulation of plant growth. Hence, rapid, tight, and coordinated regulation of its concentration is achieved by auxin modulation at multiple levels. Beyond the structural enzymes involved in auxin biosynthesis, transport, and signal transduction, transcription factors (TFs) can finely and rapidly drive auxin response in specific tissues. Auxin Response Factors (ARFs) such as the ARF4, 7, 8, 19 and many other TF families, such as WRKY and MADS, have been identified to play a role in modulating various auxin-mediated responses in recent times. Here, we review the most relevant and recent literature on TFs associated with the regulation of the biosynthetic, transport, and signalling auxin pathways and miRNA-related feedback loops in response to major abiotic stresses. Knowledge of the specific role of TFs may be of utmost importance in counteracting the effects of GCC on future agriculture and may pave the way for increased plant resilience."
Authors: Jorge Hernández-García, Antonio Serrano-Mislata, María Lozano-Quiles, Cristina Úrbez, María A. Nohales, Noel Blanco-Touriñán, Huadong Peng, Rodrigo Ledesma-Amaro and Miguel A. Blázquez.
PNAS (2024)
Significance: DELLA proteins are plant-specific transcriptional hubs integrating environmental signals with endogenous cues. In order to regulate downstream processes, DELLAs modulate the activity of hundreds of transcription factors (TFs) and transcriptional regulators in various ways. Here, we describe the molecular mechanism underlying DELLA coactivator function. We show that DELLAs act as transcriptional activators by interacting with the Mediator complex subunit MED15. This interaction is necessary to regulate a specific subset of DELLA-dependent responses that are mediated by transcriptional coactivation, but not those regulated by TF sequestration. We further show that this mechanism is present in bryophyte DELLAs and thus represents a conserved mechanism of DELLA function in land plants.
Abstract: "DELLA proteins are negative regulators of the gibberellin response pathway in angiosperms, acting as central hubs that interact with hundreds of transcription factors (TFs) and regulators to modulate their activities. While the mechanism of TF sequestration by DELLAs to prevent DNA binding to downstream targets has been extensively documented, the mechanism that allows them to act as coactivators remains to be understood. Here, we demonstrate that DELLAs directly recruit the Mediator complex to specific loci in Arabidopsis, facilitating transcription. This recruitment involves DELLA amino-terminal domain and the conserved MED15 KIX domain. Accordingly, partial loss of MED15 function mainly disrupted processes known to rely on DELLA coactivation capacity, including cytokinin-dependent regulation of meristem function and skotomorphogenic response, gibberellin metabolism feedback, and flavonol production. We have also found that the single DELLA protein in the liverwort Marchantia polymorpha is capable of recruiting MpMED15 subunits, contributing to transcriptional coactivation. The conservation of Mediator-dependent transcriptional coactivation by DELLA between Arabidopsis and Marchantia implies that this mechanism is intrinsic to the emergence of DELLA in the last common ancestor of land plants."
Authors: Mahamud Hossain Al-Mamun, Christopher Ian Cazzonelli and Priti Krishna.
Frontiers in Plant Science (2024)
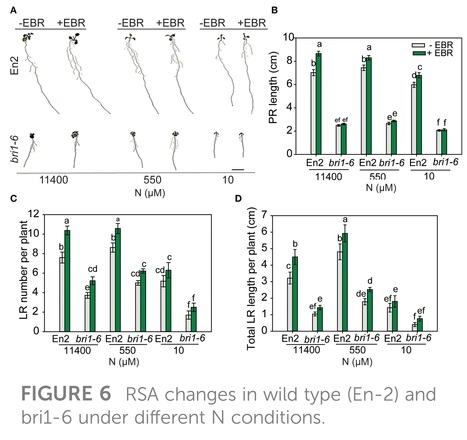
Abstract: "Plants modify their root system architecture (RSA) in response to nitrogen (N) deficiency. The plant steroidal hormone, brassinosteroid (BR), plays important roles in root growth and development. This study demonstrates that optimal levels of exogenous BR impact significant increases in lateral root length and numbers in Arabidopsis seedlings under mild N-deficient conditions as compared to untreated seedlings. The impact of BR on RSA was stronger under mild N deficiency than under N-sufficient conditions. The BR effects on RSA were mimicked in dominant mutants of BZR1 and BES1 (bzr1-1D and bes1-D) transcription factors, while the RSA was highly reduced in the BR-insensitive mutant bri1-6, confirming that BR signaling is essential for the development of RSA under both N-sufficient and N-deficient conditions. Exogenous BR and constitutive activity of BZR1 and BES1 in dominant mutants led to enhanced root meristem, meristematic cell number, and cortical cell length. Under mild N deficiency, bzr1-1D displayed higher fresh and dry shoot weights, chlorophyll content, and N levels in the shoot, as compared to the wild type. These results indicate that BR modulates RSA under both N-sufficient and N-deficient conditions via the transcription factors BES1/BZR1 module and confers tolerance to N deficiency."
Authors: Tao Wang, Xuemin Ma, Ying Chen, Cuicui Wang, Zhenxiao Xia, Zixi Liu, Lihong Gao and Wenna Zhang.
Plant, Cell & Environment (2024)
Abstract: "Low temperature stress poses a significant challenge to the productivity of horticultural crops. The dynamic expression of cold-responsive genes plays a crucial role in plant cold tolerance. While NAC transcription factors have been extensively studied in plant growth and development, their involvement in regulating plant cold tolerance remains poorly understood. In this study, we focused on the identification and characterisation of SlNAC3 as the most rapid and robust responsive gene in tomato under low temperature conditions. Manipulating SlNAC3 through overexpression or silencing resulted in reduced or enhanced cold tolerance, respectively. Surprisingly, we discovered a negative correlation between the expression of CBF and cold tolerance in the SlNAC3 transgenic lines. These findings suggest that SlNAC3 regulates tomato cold tolerance likely through a CBF-independent pathway. Furthermore, we conducted additional investigations to identify the molecular mechanisms underlying SlNAC3-mediated cold tolerance in tomatoes. Our results revealed that SlNAC3 controls the transcription of ethylene biosynthetic genes, thereby bursting ethylene release in response to cold stress. Indeed, the silencing of these genes led to an augmentation in cold tolerance. This discovery provides valuable insights into the regulatory pathways involved in ethylene-mediated cold tolerance in tomatoes, offering potential strategies for developing innovative approaches to enhance cold stress resilience in this economically important crop species."
Authors: Lei Liang and Xiangyang Hu.
Phyton (2024)
Abstract: "With the rapid development of modern molecular biology and bioinformatics, many studies have proved that transcription factors play an important role in regulating the growth and development of plants. SPATULA (SPT) belongs to the bHLH transcription family and participates in many processes of regulating plant growth and development. This review systemically summarizes the multiple roles of SPT in plant growth, development, and stress response, including seed germination, flowering, leaf size, carpel development, and root elongation, which is helpful for us to better understand the functions of SPT."
Authors: Shammi Akter, Oscar Castaneda-Méndez and Jesús Beltrán
Current Opinion in Biotechnology (2024)
Highlights • Several challenges remain for the widespread use of synthetic biology in crops. • Synthetic gene expression circuitry is advancing in model plants. • Rapid enzyme discovery and testing are needed for plant metabolic engineering.
Abstract: "Plant synthetic biology (Plant SynBio) is an emerging field with the potential to enhance agriculture, human health, and sustainability. Integrating genetic tools and engineering principles, Plant SynBio aims to manipulate cellular functions and construct novel biochemical pathways to develop plants with new phenotypic traits, enhanced yield, and be able to produce natural products and pharmaceuticals. This review compiles research efforts in reprogramming plant developmental and biochemical pathways. We highlight studies leveraging new gene expression toolkits to alter plant architecture for improved performance in model and crop systems and to produce useful metabolites in plant tissues. Furthermore, we provide insights into the challenges and opportunities associated with the adoption of Plant SynBio in addressing complex issues impacting agriculture and human health."
Authors: Xuehuan Feng, Jinfang Zheng, Iker Irisarri, Huihui Yu, Bo Zheng, Zahin Ali, Sophie de Vries, Jean Keller, Janine M. R. Fürst-Jansen, Armin Dadras, Jaccoline M. S. Zegers, Tim P. Rieseberg, Amra Dhabalia Ashok, Tatyana Darienko, Maaike J. Bierenbroodspot, Lydia Gramzow, Romy Petroll, Fabian B. Haas, Noe Fernandez-Pozo, Orestis Nousias, Tang Li, Elisabeth Fitzek, W. Scott Grayburn, Nina Rittmeier, Charlotte Permann, Florian Rümpler, John M. Archibald, Günter Theißen, Jeffrey P. Mower, Maike Lorenz, Henrik Buschmann, Klaus von Schwartzenberg, Lori Boston, Richard D. Hayes, Chris Daum, Kerrie Barry, Igor V. Grigoriev, Xiyin Wang, Fay-Wei Li, Stefan A. Rensing, Julius Ben Ari, Noa Keren, Assaf Mosquna, Andreas Holzinger, Pierre-Marc Delaux, Chi Zhang, Jinling Huang, Marek Mutwil, Jan de Vries and Yanbin Yin
Nature Genetics (2024)
Editor's view: Genome assemblies of four filamentous Zygnematophyceae and co-expression network analyses shed light on the evolutionary roots of the mechanism for balancing environmental responses and multicellular growth.
Abstract: "Zygnematophyceae are the algal sisters of land plants. Here we sequenced four genomes of filamentous Zygnematophyceae, including chromosome-scale assemblies for three strains of Zygnema circumcarinatum. We inferred traits in the ancestor of Zygnematophyceae and land plants that might have ushered in the conquest of land by plants: expanded genes for signaling cascades, environmental response, and multicellular growth. Zygnematophyceae and land plants share all the major enzymes for cell wall synthesis and remodifications, and gene gains shaped this toolkit. Co-expression network analyses uncover gene cohorts that unite environmental signaling with multicellular developmental programs. Our data shed light on a molecular chassis that balances environmental response and growth modulation across more than 600 million years of streptophyte evolution."
Authors: Yiran Xu, Shengdong Qi, Yong Wang and Jingbo Jia.
Journal of Experimental Botany (2024)
One-sentence summary: This review summarizes the interplay between nitrate and abscisic acid and provides research perspectives for breeding high nitrogen use efficiency and stress-tolerant crop varieties.
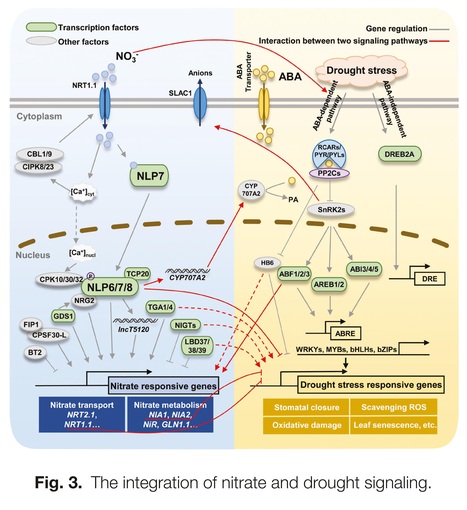
Abstract: "To meet the demands of the new Green Revolution and sustainable agriculture, it is important to develop crop varieties with improved yield, nitrogen use efficiency, and stress resistance. Nitrate is the major form of inorganic nitrogen available for plant growth in many well-aerated agricultural soils, and acts as a signaling molecule regulating plant development, growth, and stress responses. Abscisic acid (ABA), an important phytohormone, plays vital roles in integrating extrinsic and intrinsic responses and mediating plant growth and development in response to biotic and abiotic stresses. Therefore, elucidating the interplay between nitrate and ABA can contribute to crop breeding and sustainable agriculture. Here, we review studies that have investigated the interplay between nitrate and ABA in root growth modulation, nitrate and ABA transport processes, seed germination regulation, and drought responses. We also focus on nitrate and ABA interplay in several reported omics analyses with some important nodes in the crosstalk between nitrate and ABA. Through these insights, we proposed some research perspectives that could help to develop crop varieties adapted to a changing environment and to improve crop yield with high nitrogen use efficiency and strong stress resistance."
Authors: Shaoqin Li, Li He, Yongping Yang, Yixin Zhang, Xiao Han, Yanru Hu and Yanjuan Jiang.
The Plant Cell (2024)
One-sentence summary: The NPR1–TGA3–ICE1 regulatory module represents an important step in salicylic acid signaling during cold-activated resistance of plants to pathogen attack.
Abstract: "Cold stress affects plant immune responses, and this process may involve the salicylic acid (SA) signaling pathway. However, the underlying mechanism by which low-temperature signals coordinate with SA signaling to regulate plant immunity remains unclear. Here, we found that low temperatures enhanced the disease resistance of Arabidopsis thaliana against Pseudomonas syringae pv. tomato DC3000. This process required INDUCER OF CBF EXPRESSION 1 (ICE1), the core transcription factor in cold-signal cascades. ICE1 physically interacted with NONEXPRESSER OF PATHOGENESIS-RELATED GENES 1 (NPR1), the master regulator of the SA signaling pathway. Enrichment of ICE1 on the PATHOGENESIS-RELATED GENE 1 (PR1) promoter and its ability to transcriptionally activate PR1 were enhanced by NPR1. Further analyses revealed that cold stress signals cooperate with SA signals to facilitate plant immunity against pathogen attack in an ICE1-dependent manner. Cold treatment promoted interactions of NPR1 and TGACG-BINDING FACTOR 3 (TGA3) with ICE1 and increased the ability of the ICE1–TGA3 complex to transcriptionally activate PR1. Together, our results characterize a critical role of ICE1 as an indispensable regulatory node linking low-temperature-activated and SA-regulated immunity. Understanding this crucial role of ICE1 in coordinating multiple signals associated with immunity broadens our understanding of plant–pathogen interactions."
|


 Your new post is loading...
Your new post is loading...