Your new post is loading...
 Your new post is loading...
Authors: Xing Wang, Zhaobin Ren, Shipeng Xie, Zhaohu Li, Yuyi Zhou and Liusheng Duan.
Plant Physiology (2024)
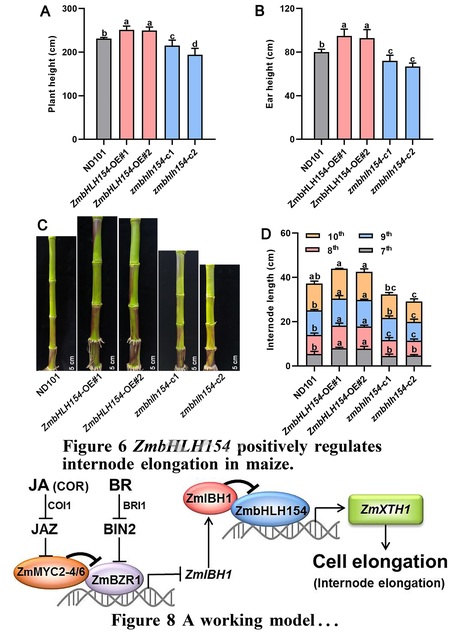
One-sentence summary: A jasmonate mimic regulates a basic helix-loop-helix network by attenuating brassinosteroid signaling, which represses expression of a cell wall–related gene and inhibits internode elongation in maize.
Abstract: "Lodging restricts growth, development, and yield formation in maize (Zea mays L.). Shorter internode length is beneficial for lodging tolerance. However, although brassinosteroids (BRs) and jasmonic acid (JA) are known to antagonistically regulate internode growth, the underlying molecular mechanism is still unclear. In this study, application of the JA mimic coronatine (COR) inhibited basal internode elongation at the jointing stage and repressed expression of the cell wall-related gene XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 1 (ZmXTH1), whose overexpression in maize plants promotes internode elongation. We demonstrated that the basic helix–loop–helix (bHLH) transcription factor ZmbHLH154 binds directly to the ZmXTH1 promoter and induces its expression, whereas the bHLH transcription factor ILI1 BINDING BHLH 1 (ZmIBH1) inhibits this transcriptional activation by forming a heterodimer with ZmbHLH154. Overexpressing ZmbHLH154 led to longer internodes, whereas zmbhlh154 mutants had shorter internodes than the wild type. The core JA-dependent transcription factors ZmMYC2-4 and ZmMYC2-6 interacted with BRASSINAZOLE RESISTANT 1 (ZmBZR1), a key factor in BR signaling, and these interactions eliminated the inhibitory effect of ZmBZR1 on its downstream gene ZmIBH1. Collectively, these results reveal a signaling module in which JA regulates a bHLH network by attenuating BR signaling to inhibit ZmXTH1 expression, thereby regulating cell elongation in maize."
Authors: Yangang Pei, Qihan Xue, Peng Shu, Weijie Xu, Xiaofei Du, Mengbo Wu, Kaidong Liu, Julien Pirrello, Mondher Bouzayen, Yiguo Hong and Mingchun Liu.
Developmental Cell (2024)
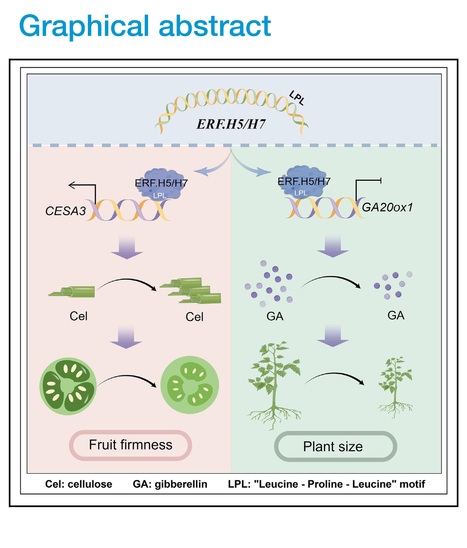
Editor's view: Pei et al. demonstrate the role of bifunctional transcription factors SlERF.H5 and SlERF.H7 in activating the cellulose biosynthesis gene SlCESA3 while repressing the gibberellin biosynthesis gene GA20ox1. The study highlights the distinct transcriptional regulatory functions of these ERFs in promoting cell wall formation and inhibiting plant growth.
Highlights • Cellulose is required for cell wall formation and firmness maintenance of fruits • SlERF.H5 and SlERF.H7 act as both transcriptional activators and repressors • The regulatory activity of SlERF.H5 and SlERF.H7 is mediated by a conserved motif
Abstract: "The plant cell wall is a dynamic structure that plays an essential role in development, but the mechanism regulating cell wall formation remains poorly understood. We demonstrate that two transcription factors, SlERF.H5 and SlERF.H7, control cell wall formation and tomato fruit firmness in an additive manner. Knockout of SlERF.H5, SlERF.H7, or both genes decreased cell wall thickness, firmness, and cellulose contents in fruits during early development, especially in double-knockout lines. Overexpressing either gene resulted in thicker cell walls and greater fruit firmness with elevated cellulose levels in fruits but severely dwarf plants with lower gibberellin contents. We further identified that SlERF.H5 and SlERF.H7 activate the cellulose biosynthesis gene SlCESA3 but repress the gibberellin biosynthesis gene GA20ox1. Moreover, we identified a conserved LPL motif in these ERFs responsible for their activities as transcriptional activators and repressors, providing insight into how bifunctional transcription factors modulate distinct developmental processes."
Authors: Wei Li, Ying-Chung Jimmy Lin, Ying-Lan Chen, Chenguang Zhou, Shuang Li, Nette De Ridder, Dyoni M. Oliveira, Lanjun Zhang, Baocai Zhang, Jack P. Wang, Changzheng Xu, Xiaokang Fu, Keming Luo, Ai-Min Wu, Taku Demura, Meng-Zhu Lu, Yihua Zhou, Laigeng Li, Toshiaki Umezawa, Wout Boerjan and Vincent L. Chiang.
Molecular Plant (2024)
Abstract: "Cell walls in plants, particularly forest trees, are the major carbon sink of the terrestrial ecosystem. Chemical and biosynthetic features of plant cell walls were revealed early on, focusing mostly on herbaceous model species. Recent developments in genomics, transcriptomics, epigenomics, transgenesis, and associated analytical techniques are enabling novel insights into formation of woody cell walls. Here, we review multilevel regulation of cell wall biosynthesis in forest tree species. We highlight current approaches to engineering cell walls as potential feedstock for materials and energy and survey reported field tests of such engineered transgenic trees. We outline opportunities and challenges in future research to better understand cell type biogenesis for more efficient wood cell wall modification and utilization for biomaterials or for enhanced carbon capture and storage."
Authors: Ruo-Xi Zhang, Yudi Liu, Xian Zhang, Xiaomei Chen, Juanli Sun, Yun Zhao, Jinyun Zhang, Jia-Long Yao, Liao Liao, Hui Zhou and Yuepeng Han.
New Phytologist (2024)
Abstract: "Although maturity date (MD) is an essential factor affecting fresh fruit marketing and has a pleiotropic effect on fruit taste qualities, the underlying mechanisms remain largely unclear. In this study, we functionally characterized two adjacent NAM-ATAF1/2-CUC2 (NAC) transcription factors (TFs), PpNAC1 and PpNAC5, both of which were associated with fruit MD in peach. PpNAC1 and PpNAC5 were found capable of activating transcription of genes associated with cell elongation, cell wall degradation and ethylene biosynthesis, suggesting their regulatory roles in fruit enlargement and ripening. Furthermore, PpNAC1 and PpNAC5 had pleiotropic effects on fruit taste due to their ability to activate transcription of genes for sugar accumulation and organic acid degradation. Interestingly, both PpNAC1 and PpNAC5 orthologues were found in fruit-producing angiosperms and adjacently arranged in all 91 tested dicots but absent in fruitless gymnosperms, suggesting their important roles in fruit development. Our results provide insight into the regulatory roles of NAC TFs in MD and fruit taste."
Authors: Shuhuan Zhang, Weihao Miao, Ye Liu, Jiafu Jiang, Sumei Chen, Fadi Chen and Zhiyong Guan.
BMC Genomics (2023)
Abstract: "Background - Black spot disease caused by the necrotrophic fungus Alternaria spp. is one of the most devastating diseases affecting Chrysanthemum morifolium. There is currently no effective way to prevent chrysanthemum black spot. Results - We revealed that pre-treatment of chrysanthemum leaves with the methyl jasmonate (MeJA) significantly reduces their susceptibility to Alternaria alternata. To understand how MeJA treatment induces resistance, we monitored the dynamics of metabolites and the transcriptome in leaves after MeJA treatment following A. alternata infection. JA signaling affected the resistance of plants to pathogens through cell wall modification, Ca2+ regulation, reactive oxygen species (ROS) regulation, mitogen‐activated protein kinase cascade and hormonal signaling processes, and the accumulation of anti-fungal and anti-oxidant metabolites. Furthermore, the expression of genes associated with these functions was verified by reverse transcription quantitative PCR and transgenic assays. Conclusion - Our findings indicate that MeJA pre-treatment could be a potential orchestrator of a broad-spectrum defense response that may help establish an ecologically friendly pest control strategy and offer a promising way of priming plants to induce defense responses against A. alternata."
Authors: Vojtech Didi, Dominique Arnaud, Anna Pacinkova, Radek Jupa, Radim Cegan, Alesia Melnikava, Jana Vasickova, Mariana Benitez, Faride Unda, Tereza Dobisova, Willi Riber, Zuzana Dostalova, Shawn D. Mansfield, Ondrej Novak, Miroslav Strnad, Roman Hobza, Vit Gloser, Eva Budinska and Jan Hejatko.
bioRxiv (2023)
Abstract: "Spatiotemporal control over developmental programs is vital to all organisms. Here we show that cytokinin (signaling) deficiency leads to early secondary cell wall (SCW) formation in Arabidopsis inflorescence stem that associates with precocious upregulation of a SCW transcriptional cascade controlled by NAC TFs (NSTs). We demonstrate that cytokinin signaling through the AHK2/3 and the ARR1/10/12 suppresses the expression of several NSTs and SCW formation in the apical portions of stems. Exogenous cytokinin application reconstituted both proper development and apical-basal gradient of NST1 and NST3 in a cytokinin biosynthesis-deficient mutant. We show that AHK2 and AHK3 required functional NST1 or NST3 to control SCW initiation in the interfascicular fibers, further evidencing that cytokinins act upstream of NSTs transcription factors. The premature onset of a rigid SCW biosynthesis and altered expression of NST1/3 and VND6/7 due to cytokinin deficiency led to the formation of smaller tracheary elements (TEs) and impaired hydraulic conductivity. We conclude that cytokinins downregulate NSTs to inhibit premature SCW formation in the apical part of the inflorescence stem, facilitating thus the development of fully functional TEs and interfascicular fibers."
Authors: Yun Wei, Zhi Liu, Tianxing Lv, Yaxiu Xu, Yajing Wei, Weiting Liu, Li Liu, Aide Wang and Tong Li.
The Plant Cell (2023)
Abstract: "The phytohormone ethylene plays an important role in promoting the softening of climacteric fruits, such as apples (Malus domestica); however, important aspects of the underlying regulatory mechanisms are not well understood. In this study, we identified apple MITOGEN-ACTIVATED PROTEIN KINASE 3 (MdMAPK3) as an important positive regulator of ethylene-induced apple fruit softening during storage. Specifically, we show that MdMAPK3 interacts with and phosphorylates the transcription factor NAM-ATAF1/2-CUC2 72 (MdNAC72), which functions as a transcriptional repressor of the cell wall degradation-related gene POLYGALACTURONASE1 (MdPG1). The increase in MdMAPK3 kinase activity was induced by ethylene, which promoted the phosphorylation of MdNAC72 by MdMAPK3. Additionally, MdPUB24 functions as an E3 ubiquitin ligase to ubiquitinate MdNAC72, resulting in its degradation via the 26S proteasome pathway, which was enhanced by ethylene-induced phosphorylation of MdNAC72 by MdMAPK3. The degradation of MdNAC72 increased the expression of MdPG1, which in turn promoted apple fruit softening. Notably, using variants of MdNAC72 that were mutated at specific phosphorylation sites, we observed that the phosphorylation state of MdNAC72 affected apple fruit softening during storage. This study thus reveals that the ethylene-MdMAPK3-MdNAC72-MdPUB24 module is involved in ethylene-induced apple fruit softening, providing insights into climacteric fruit softening."
Authors: Yang-Shuo Dai, Di Liu, Wuxiu Guo, Zhi-Xuan Liu, Xue Zhang, Li-Li Shi, De-Mian Zhou, Ling-Na Wang, Kui Kang, Feng-Zhu Wang, Shan-Shan Zhao, Yi-Fang Tan, Tian Hu, Wu Chen, Peng Li, Qing-Ming Zhou, Long-Yu Yuan, Zhenfei Zhang, Yue-Qin Chen, Wen-Qing Zhang, Juan Li, Lu-Jun Yu and Shi Xiao.
Plant Biotechnology Journal (2023)
Abstract: "Brown planthopper (BPH, Nilaparvata lugens), a highly destructive insect pest, poses a serious threat to rice (Oryza sativa) production worldwide. Jasmonates are key phytohormones that regulate plant defences against BPH; however, the molecular link between jasmonates and BPH responses in rice remains largely unknown. Here, we discovered a Poaceae-specific metabolite, mixed-linkage β-1,3;1,4-d-glucan (MLG), which contributes to jasmonate-mediated BPH resistance. MLG levels in rice significantly increased upon BPH attack. Overexpressing OsCslF6, which encodes a glucan synthase that catalyses MLG biosynthesis, significantly enhanced BPH resistance and cell wall thickness in vascular bundles, whereas knockout of OsCslF6 reduced BPH resistance and vascular wall thickness. OsMYC2, a master transcription factor of jasmonate signalling, directly controlled the upregulation of OsCslF6 in response to BPH feeding. The AT-rich domain of the OsCslF6 promoter varies in rice varieties from different locations and natural variants in this domain were associated with BPH resistance. MLG-derived oligosaccharides bound to the plasma membrane-anchored LECTIN RECEPTOR KINASE1 OsLecRK1 and modulated its activity. Thus, our findings suggest that the OsMYC2-OsCslF6 module regulates pest resistance by modulating MLG production to enhance vascular wall thickness and OsLecRK1-mediated defence signalling during rice-BPH interactions."
Authors: Maria C. Camarero, Beatriz Briegas, Jorge Corbacho, Juana Labrador, Mercedes Gallardo and Maria C. Gomez-Jimenez.
International Journal of Molecular Sciences (2023)
Abstract: "In the olive (Olea europaea L.), an economically leading oil crop worldwide, fruit size and yield are determined by the early stages of fruit development. However, few detailed analyses of this stage of fruit development are available. This study offers an extensive characterization of the various processes involved in early olive fruit growth (cell division, cell cycle regulation, and cell expansion). For this, cytological, hormonal, and transcriptional changes characterizing the phases of early fruit development were analyzed in olive fruit of the cv. ‘Picual’. First, the surface area and mitotic activity (by flow cytometry) of fruit cells were investigated during early olive fruit development, from 0 to 42 days post-anthesis (DPA). The results demonstrate that the cell division phase extends up to 21 DPA, during which the maximal proportion of 4C cells in olive fruits was reached at 14 DPA, indicating that intensive cell division was activated in olive fruits at that time. Subsequently, fruit cell expansion lasted as long as 3 weeks more before endocarp lignification. Finally, the molecular mechanisms controlling the early fruit development were investigated by analyzing the transcriptome of olive flowers at anthesis (fruit set) as well as olive fruits at 14 DPA (cell division phase) and at 28 DPA (cell expansion phase). Sequential induction of the cell cycle regulating genes is associated with the upregulation of genes involved in cell wall remodeling and ion fluxes, and with a shift in plant hormone metabolism and signaling genes during early olive fruit development. This occurs together with transcriptional activity of subtilisin-like protease proteins together with transcription factors potentially involved in early fruit growth signaling. This gene expression profile, together with hormonal regulators, offers new insights for understanding the processes that regulate cell division and expansion, and ultimately fruit yield and olive size."
Authors: Zhenzhen Peng, Gangshuai Liu, Hongli Li, Yunxiang Wang, Haiyan Gao, Tomislav Jemrić and Daqi Fu.
International Journal of Molecular Sciences (2022)
Abstract: "Fruit softening that occurs during fruit ripening and postharvest storage determines the fruit quality, shelf life and commercial value and makes fruits more attractive for seed dispersal. In addition, over-softening results in fruit eventual decay, render fruit susceptible to invasion by opportunistic pathogens. Many studies have been conducted to reveal how fruit softens and how to control softening. However, softening is a complex and delicate life process, including physiological, biochemical and metabolic changes, which are closely related to each other and are affected by environmental conditions such as temperature, humidity and light. In this review, the current knowledge regarding fruit softening mechanisms is summarized from cell wall metabolism (cell wall structure changes and cell-wall-degrading enzymes), plant hormones (ETH, ABA, IAA and BR et al.), transcription factors (MADS-Box, AP2/ERF, NAC, MYB and BZR) and epigenetics (DNA methylation, histone demethylation and histone acetylation) and a diagram of the regulatory relationship between these factors is provided. It will provide reference for the cultivation of anti-softening fruits."
Authors: Yuting Meng, Jing Huang, Huaikang Jing, Qi Wu, Renfang Shen and Xiaofang Zhu.
Plant Science (2022)
Highlights: • Exogenous ABA decreased the fixation of Cd in root cell wall hemicelluloses. • Exogenous ABA down-regulated the expression of genes related to Cd uptake and translocation. • Exogenous ABA alleviates Cd toxicity by promoting Cd efflux and cytoplasmic chelation rather than vacuolar chelation. • ABI4 might negatively regulate IRT1, ZIP1, ZIP4, HMA2 and HMA4, positively regulate PDF2.6 and PDR8.
Abstract: "Exogenous abscisic acid (ABA) has been implicated in plant response to cadmium (Cd) stress, but the underlying mechanism remains unclear. In the present study, we found that exogenous ABA application decreased Cd fixation in wild type (WT) root cell wall through reducing the hemicelluloses content, in parallel with the decreased expression of IRT1, ZIP1, ZIP4, HMA2 and HMA4, which are related to Cd uptake and translocation, and the increased expression of PDF2.6, PDR8 and AIT1, which are related to Cd chelation, efflux, and accumulation inhibition. These changes might be associated with the reduced Cd accumulation in roots and shoots and the alleviated Cd toxicity. In contrast, the mutation of ABI4, a transcription factor in ABA signaling pathway, significantly increased the expression of IRT1, ZIP1, ZIP4, HMA2 and HMA4, while decreased the expression of AIT1, PDF2.6 and PDR8, enhancing Cd accumulation in roots and shoots of abi4. The enhanced Cd-sensitivity in abi4 mutant could not be rescued by exogenous ABA addition compared with WT. In a word, we conclude that exogenous ABA mitigates Cd toxicity in Arabidopsis thaliana via inhibiting Cd uptake, translocation and accumulation, promoting Cd chelation and efflux, a pathway that might be regulated by ABI4."
Authors: Yang Zhang, Yingying Liu, Xueying Wang, Ruiqi Wang, Xuebing Chen, Shuang Wang, Hairong Wei and Zhigang Wei.
Frontiers in Plant Science (2022)
Abstract: "WUSCHEL-related homeobox (WOX) genes are plant-specific transcription factors (TFs) involved in multiple processes of plant development. However, there have hitherto no studies on the WOX TFs involved in secondary cell wall (SCW) formation been reported. In this study, we identified a Populus trichocarpa WOX gene, PtrWOX13A, which was predominantly expressed in SCW, and then characterized its functions through generating PtrWOX13A overexpression poplar transgenic lines; these lines exhibited not only significantly enhanced growth potential, but also remarkably increased SCW thicknesses, fiber lengths, and lignin and hemicellulose contents. However, no obvious change in cellulose content was observed. We revealed that PtrWOX13A directly activated its target genes through binding to two cis-elements, ATTGATTG and TTAATSS, in their promoter regions. The fact that PtrWOX13A responded to the exogenous GAs implies that it is responsive to GA homeostasis caused by GA inactivation and activation genes (e.g., PtrGA20ox4, PtrGA2ox1, and PtrGA3ox1), which were regulated by PtrWOX13A directly or indirectly. Since the master switch gene of SCW formation, PtrWND6A, and lignin biosynthesis regulator, MYB28, significantly increased in PtrWOX13A transgenic lines, we proposed that PtrWOX13A, as a higher hierarchy TF, participated in SCW formation through controlling the genes that are components of the known hierarchical transcription regulation network of poplar SCW formation, and simultaneously triggering a gibberellin-mediated signaling cascade. The discovery of PtrWOX13A predominantly expressed in SCW and its regulatory functions in the poplar wood formation has important implications for improving the wood quality of trees via genetic engineering."
|
Authors: Zhichao Sun, Xinmiao Guo, R.M. Saravana Kumar, Chunying Huang, Yan Xie, Meng Li and Jisheng Li.
Plant Science (2024)
Highlights: • Late stage of mulberry fruit ripening witnessed ethylene accumulation and physiological changes. • Late stage fruit ripening-specific differentially expressed genes and associated metabolic pathways highlighted. • Transcriptome and metabolome data interconnects ethylene signaling and mulberry fruit ripening. • MaERF3 are important for ripening process.
Abstract: "Mulberry (Morus alba L.) is a climacteric and highly perishable fruit. Ethylene has been considered to be an important trigger of fruit ripening process. However, the role of ethylene in the mulberry fruit ripening process remains unclear. In this study, we performed a comprehensive analysis of metabolomic and transcriptomic data of mulberry fruit and the physiological changes accompanying the fruit ripening process. Our study revealed that changes in the accumulation of specific metabolites at different stages of fruit development and ripening were closely correlated to transcriptional changes as well as underlying physiological changes and the development of taste biomolecules. The ripening of mulberry fruits was highly associated with the production of endogenous ethylene, and further application of exogenous ethylene assisted the ripening process. Transcriptomic analysis revealed that differential expression of diverse ripening-related genes was involved in sugar metabolism, anthocyanin biosynthesis, and cell wall modification pathways. Network analysis of transcriptomics and metabolomics data revealed that many transcription factors and ripening-related genes were involved, among which ethylene-responsive transcription factor 3 (MaERF3) plays a crucial role in the ripening process. The role of MaERF3 in ripening was experimentally proven in a transient overexpression assay in apples. Our study indicates that ethylene plays a vital role in modulating mulberry fruit ripening. The results provide a basis for guiding the genetic manipulation of mulberry fruits towards sustainable agricultural practices and improve post-harvest management, potentially enhancing the quality and shelf life of mulberry fruits for sustainable agriculture and forestry."
Authors: James Giovannoni, Yao Chen, Xin Wang, Vincent Colantonio, Tara Fish, Jie Ye, Theodore Thannhauser, Zhibiao Ye, Mingchun Liu, Yongsheng Liu and Zhangjun Fei.
Research Square (2024)
Abstract: "Ripening is crucial for the development of fleshy fruits that release their seeds following consumption by frugivores and are important contributors to human health and nutritional security. Many genetic ripening regulators have been identified, especially in the model system tomato, yet more remain to be discovered and integrated into comprehensive regulatory models. Most tomato ripening genes have been studied in pericarp tissue, though recent evidence indicates that locule tissue is a site of early ripening-gene activities. Here we identified and functionally characterized an Ethylene Response Factor gene, SlERF.D6, by investigating tomato transcriptome data throughout plant development, emphasizing genes elevated in the locule during fruit development and ripening. SlERF.D6loss-of-function mutants resulting from CRISPR/Cas9 gene editing delayed ripening initiation and carotenoid accumulation in both pericarp and locule tissues. Transcriptome analysis of lines altered in SlERF.D6 expression revealed multiple classes of altered genes including ripening regulators, in addition to carotenoid, cell wall and ethylene pathway genes, suggesting comprehensive ripening control. Distinct regulatory patterns in pericarp versus locule tissues were observed indicating tissue-specific activity of this transcription factor. Analysis of SlERF.D6 interaction with target promoters revealed an AP2/ERF transcription factor (SlDEAR2) as a target of SlERF.D6. Furthermore, we show that a third transcription factor gene, SlTCP12, is a target of SlDEAR2, presenting a tri-component module of ripening control."
Authors: Jong Hee Im, Seungmin Son, Won-Chan Kim, Kihwan Kim, Nobutaka Mitsuda, Jae-Heung Ko and Kyung-Hwan Han.
The Plant Journal (2024)
Abstract: "Formation of secondary cell wall (SCW) is tightly regulated spatiotemporally by various developmental and environmental signals. Successful fine-tuning of the trade-off between SCW biosynthesis and stress responses requires a better understanding of how plant growth is regulated under environmental stress conditions. However, the current understanding of the interplay between environmental signaling and SCW formation is limited. The lipid-derived plant hormone jasmonate (JA) and its derivatives are important signaling components involved in various physiological processes including plant growth, development, and abiotic/biotic stress responses. Recent studies suggest that JA is involved in SCW formation but the signaling pathway has not been studied for how JA regulates SCW formation. We tested this hypothesis using the transcription factor MYB46, a master switch for SCW biosynthesis, and JA treatments. Both the transcript and protein levels of MYB46, a master switch for SCW formation, were significantly increased by JA treatment, resulting in the upregulation of SCW biosynthesis. We then show that this JA-induced upregulation of MYB46 is mediated by MYC2, a central regulator of JA signaling, which binds to the promoter of MYB46. We conclude that this MYC2-MYB46 module is a key component of the plant response to JA in SCW formation."
Authors: Keming Luo, Shuai Liu, Xiaokang Fu, Xuelian Du, Jian Hu, Lianjia Luo, Changjian Fa, Rongling Wu, Laigeng Li and Changzheng Xu.
Research Square (2023)
Abstract: "Auxin, as a vital phytohormone, is enriched in the vascular cambium, playing a crucial role in regulating wood formation in trees. Despite its significance, the molecular mechanisms underlying the influence of auxin on wood development remain elusive. In this study, we report a transcription factor, PLETHORA 5 (PLT5), whose expression was specifically activated by auxin signalling in the vascular cambium. PLT5 was found to regulate cell expansion and lignification of fibres in poplar. Genetic experiments confirmed the noncell-autonomous regulation of auxin signalling from the vascular cambium and revealed the necessity of PLT5 protein mobility to mediate this process. Remarkably, PLT5 proteins specifically inhibit the initiation of fibre cell wall thickening by directly repressing SND1 genes. This study unveils a sophisticated model wherein the auxin-PLT5 signalling cascade intricately regulates wood fibre development in poplar by fine-tuning the thickening of fibre cell walls."
Authors: Wenyi Wang, Vanika Garg, Rajeev K. Varshney and Hao Liu.
Trends in Plant Science (2023).
Abstract: Phytohormone signaling regulates plant growth and development. Single cell RNA sequencing (scRNA-seq) provides unprecedented opportunities to decipher hormone-mediated spatiotemporal gene regulatory networks. In a recent study, Nolan et al. used time-series scRNA-seq to identify the cortex as a key site for brassinosteroid (BR)-mediated gene expression and revealed a signaling network during cell phase transition."
Authors: Pingxia Zhao, Jing Zhang, Siyan Chen, Zisheng Zhang, Guangyu Wan, Jieli Mao, Zhen Wang, Shutang Tan and Chengbin Xiang.
Cell Reports (2023)
Editor's view: Zhao et al. show that ERF1, a hub transcription factor in the stress response, inhibits lateral root emergence by promoting high local auxin accumulation with altered distribution in the epidermis, cortex, and endodermis overlying the lateral root primordia and regulating auxin signaling in adaptation to fluctuating environments.
Highlights • ERF1 functions as a negative regulator of lateral root emergence • ERF1 enhances auxin transport by upregulating PIN1 and AUX1 • ERF1 transcriptionally represses ARF7 • ERF1 downregulates cell-wall remodeling genes in lateral root emergence
Abstract: "Lateral roots (LRs) are crucial for plants to sense environmental signals in addition to water and nutrient absorption. Auxin is key for LR formation, but the underlying mechanisms are not fully understood. Here, we report that Arabidopsis ERF1 inhibits LR emergence by promoting local auxin accumulation with altered distribution and regulating auxin signaling. Loss of ERF1 increases LR density compared with the wild type, whereas ERF1 overexpression causes the opposite phenotype. ERF1 enhances auxin transport by upregulating PIN1 and AUX1, resulting in excessive auxin accumulation in the endodermal, cortical, and epidermal cells surrounding LR primordia. Furthermore, ERF1 represses ARF7 transcription, thereby downregulating the expression of cell-wall remodeling genes that facilitate LR emergence. Together, our study reveals that ERF1 integrates environmental signals to promote local auxin accumulation with altered distribution and repress ARF7, consequently inhibiting LR emergence in adaptation to fluctuating environments."
Authors: Shaoxue Cao, Yan Wang, Yihong Gao, Rui Xu, Jianing Ma, Zuopeng Xu, Keke Shang-Guan, Baocai Zhang and Yihua Zhou.
Molecular Plant (2023)
Abstract: "The orderly deposition of secondary cell wall (SCW) in plants is implicated in various biological programs and is precisely controlled. Although many positive and negative regulators have been documented, fine-tuning regulators of SCW formation that orchestrate distinct cellular physiologies have rarely been reported. Here, we report a SCW regulator Cellulose Synthase coexpressed Kinase1 (CSK1) and its signaling cascade in rice. Transcriptome deep sequencing of developing internodes and genome-wide co-expression assays revealed that a receptor-like cytoplasmic kinase CSK1 is co-expressed with cellulose synthase genes and is responsive to various stress stimuli. Increased SCW thickness and vigorous vessel transport of csk1 defines CSK1 as a negative regulator of SCW biosynthesis. Through observation of the signal of green fluorescent protein tagged CSK1 in rice protoplasts and stable transgenic plants, we found that CSK1 is localized in the nucleus and adjacent to the plasma membrane. Multiple lines of evidence demonstrated that CSK1 phosphorylates VASCULAR-RELATED NAC-DOMAIN 6 (VND6), a master SCW-associated transcription factor, in the nucleus, which alleviates the transcription of a suite of SCW-related genes, thereby attenuating SCW accumulation. Genetic studies validated that CSK1 functions upstream of VND6. Furthermore, physiological analyses revealed that CSK1 and VND6 implicate in abscisic acid-mediated stress signaling, which regulates cell growth and SCW deposition. The CSK1-VND6 module is necessary for the operation of SCW biosynthesis machinery, which coordinates SCW accumulation and growth plasticity. This study thus identifies a SCW fine-tuning regulator and outlines a mechanism for precise control of SCW deposition, offering tools for rationally tailoring agronomic traits."
Authors: Kyung Hwan Han, Jong Hee Im, Seungmin Son, Won-Chan Kim, Kihwan Kim, Nobutaka Mitsuda and Jae-Heung Ko.
Authorea (2023)
Abstract: "The formation of secondary cell walls is tightly regulated spatio-temporally by various developmental and environmental signals. Successful fine-tuning of the trade-off between secondary cell wall biosynthesis and stress responses requires better understanding of how plant growth is regulated under environmental stress conditions. However, current understanding of the interplay between environmental signaling and secondary cell wall formation is limited. The lipid-derived plant hormone jasmonate (JA) and its derivatives are important signaling components involved in various physiological processes including plant growth, development, and abiotic/biotic stress response. Recent studies suggest that JA may be involved in secondary cell wall formation. We tested this hypothesis using the transcription factor MYB46, a master switch for secondary wall biosynthesis, and JA treatments. Both the transcripts and protein levels of MYB46 were significantly increased by the JA treatments, which also triggered the upregulation of MYB46 downstream genes with increased secondary wall formation. We then show that this JA-induced upregulation of MYB46 function was mediated by MYC2, a basic helix-loop-helix (bHLH) domain–containing transcription factor, which plays a pivotal role in the JA-mediated changes. We conclude that this MYC2-MYB46 module is a key component of the plant response to JA signaling."
Authors: JiaQian Zhou, XiaoYang Zhao, Sen Yang, Cai E. Wu, ZhaoHui Xue and XiaoHong Kou.
Scientia Horticulturae (2023)
Highlights: • SNAC4 and SNAC9 play a significant regulatory role in tomato fruit ripening. • SNAC4/9 induced ethylene and carotenoid biosynthesis, and soluble solid content. • SNAC4 and SNAC9 exhibit different function on ABA biosynthesis and signaling. • SNAC4 and SNAC9 have different regulation modes on fruit softening. • NACs, ABA and ethylene co-regulate tomato fruit ripening.
Abstract: "Previous study suggested that SNAC4 (NM_001279348.2) and SNAC9 (NM_001365397.1) have opposite effects on carotenoid biosynthesis and softening in tomato (Solanum lycopersicum L.) fruit ripening. Here, we use an overexpressing system to explore the regulatory mechanism of SNAC4/9 in tomato fruit ripening and softening. Our results showed that the overexpression of SNAC4 and SNAC9 (OE-SNAC4/9) accelerated fruit ripening, promoted ethylene biosynthesis and carotenoid accumulation, but showed different functions on ABA content and fruit firmness. Genes related to ABA and fruit softening were up-regulated in OE-SNAC4, while down-regulated in OE-SNAC9. The structures of cell wall and peel were also different in these two OE lines. These data verified that SNAC4 and SNAC9 exhibit different regulation modes on fruit softening. Thus, we propose a working model that SNAC4/9 regulates fruit ripening and softening through three layers including cell wall modification-related genes, ethylene and ABA signaling. Our findings provide new insight into NACs involved in regulating fruit ripening."
Author: Sachihiro Matsunaga.
Molecular Plant (2022)
Excerpts: "Specifically, the malectin-like receptor kinase FERONIA (FER) is a mechanochemical sensor that monitors chemical properties through its extracellular domain at the interspace between the cell wall and the plasma membrane (Zhu et al. 2021) (Figure 1). However, the link between the signaling from the cytosolic kinase domain of FER and the transcriptional regulation in the nucleus is unknown."
"Canher et al. (2022) characterized two transcription factors that provide insights into the molecular basis of mechanotransduction. Specifically, two ETHYLENE RESPONSIVE FACTORs (ERFs), ERF114 and ERF115, mediate the gene expression induced by the perception of mechanical stress at the cell wall (Figure 1)."
"Thus, Canher et al. (2022) revealed that ERF114 and ERF115 positively regulate LR formation in roots via auxin signaling."
"The manual bending of roots decreases ERF114 expression and the formation of LR primordia following the mutation of BRASSINOSTEROID INSENSITIVE 1, which leads to a lack of BR signaling (Canher et al. 2022). This suggests that BR signaling is required for the induction of ERF114 expression, which leads to LR formation."
Authors: Balkan Canher, Fien Lanssens, Ai Zhang, Anchal Bisht, Shamik Mazumdar, Jefri Heyman, Sebastian Wolf, Charles W. Melnyk and Lieven De Veylder.
Molecular Plant (2022)
Abstract: "Plants show an unparalleled regenerative capacity, allowing them to survive severe stress conditions, such as injury, herbivory attack and harsh weather conditions. This potential not only replenishes tissues and restores damaged organs, but can also give rise to whole plant bodies. Despite the intertwined nature of development and regeneration, upstream cues and signaling mechanisms that commonly activate organogenesis are largely unknown. Here, we demonstrate that next to being activators of regeneration, ETHYLENE RESPONSE FACTOR 114 (ERF114) and ERF115 govern developmental growth in the absence of wounding or injury. Increased ERF114 and ERF115 activity enhances auxin sensitivity, which is correlated with enhanced xylem maturation and lateral root formation, whereas their knockout results in a decrease in lateral roots. Moreover, we provide evidence that mechanical cues contribute to ERF114 and ERF115 expression in correlation with BZR1-mediated brassinosteroid signaling under both regenerative and developmental conditions. Antagonistically, cell wall integrity surveillance via mechanosensory FERONIA signaling suppresses their expression under both conditions. Our data suggest a molecular framework in which cell wall signals and mechanical strains regulate organ development and regenerative responses via ERF114 and ERF115 mediated auxin signaling."
Authors: Elena Mattus-Araya, Yazmina Stappung, Raúl Herrera and María A. Moya-León.
Journal of Plant Growth Regulation (2023)
Abstract: "Fragaria chiloensis is a native species from Chile and has interesting attributes such as good taste, aroma, and exotic white color. As the maternal relative of commercial strawberry, it is interesting to understand its ripening physiology. Changes in physiological parameters (size, soluble solids, acidity, and firmness) were followed during the development and ripening of F. chiloensis fruit, noticing a fast and intense softening that severely limits its post-harvest shelf life. The content of abscisic acid (ABA) was quantified along its development and displayed an incremental pattern, meanwhile a descendant profile was determined for auxin. To clarify the role of ABA on F. chiloensis softening, detached immature fruits (C2 stage, low ABA levels) were treated with exogenous ABA and changes in firmness were followed during storage at 20 °C. Fruit firmness reduction was accelerated by ABA treatment compared to non-treated fruit, and consistent with that, a clear induction in the accumulation of transcripts of cell wall disassembly genes such as FcPG, FcRGL1, and FcExp5 was observed. In parallel, there was an increment in the transcript level of transcription factors (TFs) such as FcNAC1, FcSEP3, and FcSHP. Good correlations were determined between firmness reduction and the expression of cell wall genes and TFs. Bioinformatic analysis reveals the presence of cis elements responding to ABA in the promoter regions of cell wall disassembly genes and FcNAC1. This indicates that ABA accelerates the set of transcriptional changes leading to softening on this non-climacteric fruit and suggests that these TFs could act as molecular mediators."
|



 Your new post is loading...
Your new post is loading...