Your new post is loading...
 Your new post is loading...
Authors: Bihai Shi, Amelia Felipo-Benavent, Guillaume Cerutti, Carlos Galvan-Ampudia, Lucas Jilli, Geraldine Brunoud, Jérome Mutterer, Elody Vallet, Lali Sakvarelidze-Achard, Jean-Michel Davière, Alejandro Navarro-Galiano, Ankit Walia, Shani Lazary, Jonathan Legrand, Roy Weinstain, Alexander M. Jones, Salomé Prat, Patrick Achard and Teva Vernoux.
Nature Communications (2024)
Editor's view: Engineering of a biosensor allows the authors to map the signaling activity of the phytohormones gibberellins (GAs) and to show that GAs orient cell division at the shoot apex to establish the organization in parallel cell files of plant stems.
Abstract: "Growth at the shoot apical meristem (SAM) is essential for shoot architecture construction. The phytohormones gibberellins (GA) play a pivotal role in coordinating plant growth, but their role in the SAM remains mostly unknown. Here, we developed a ratiometric GA signaling biosensor by engineering one of the DELLA proteins, to suppress its master regulatory function in GA transcriptional responses while preserving its degradation upon GA sensing. We demonstrate that this degradation-based biosensor accurately reports on cellular changes in GA levels and perception during development. We used this biosensor to map GA signaling activity in the SAM. We show that high GA signaling is found primarily in cells located between organ primordia that are the precursors of internodes. By gain- and loss-of-function approaches, we further demonstrate that GAs regulate cell division plane orientation to establish the typical cellular organization of internodes, thus contributing to internode specification in the SAM."
Authors: Amanpreet Kaur, Norman B. Best, Thomas Hartwig, Josh Budka, Rajdeep Khangura, Steven McKenzie, Alejandro Aragón-Raygoza, Josh Strable, Burkhard Schulz and Brian P. Dilkes.
Plant Physiology (2023)
One-sentence summary: Molecular identity of a maize semidwarf mutant reveals a role for the maize GRAS domain transcription factor ortholog of DWARF AND LOW TILLERING in brassinosteroid signaling.
Abstract: "Brassinosteroids (BR) and gibberellins (GA) regulate plant height and leaf angle in maize (Zea mays). Mutants with defects in BR or GA biosynthesis or signaling identify components of these pathways and enhance our knowledge about plant growth and development. In this study, we characterized three recessive mutant alleles of GRAS transcription factor 42 (gras42) in maize, a GRAS transcription factor gene orthologous to the DWARF AND LOW TILLERING (DLT) gene of rice (Oryza sativa). These maize mutants exhibited semi-dwarf stature, shorter and wider leaves, and more upright leaf angle. Transcriptome analysis revealed a role for GRAS42 as a determinant of BR signaling. Analysis of the expression consequences from loss of GRAS42 in the gras42-mu1021149 mutant indicated a weak loss of BR signaling in the mutant, consistent with its previously demonstrated role in BR signaling in rice. Loss of BR signaling was also evident by the enhancement of weak BR biosynthetic mutant alleles in double mutants of nana plant1-1 and gras42-mu1021149. The gras42-mu1021149 mutant had little effect on GA-regulated gene expression, suggesting that GRAS42 is not a regulator of core GA signaling genes in maize. Single cell expression data identified gras42 expressed among cells in the G2/M phase of the cell cycle consistent with its previously demonstrated role in cell cycle gene expression in Arabidopsis (Arabidopsis thaliana). Cis-acting natural variation controlling GRAS42 transcript accumulation was identified by expression genome-wide association study (eGWAS) in maize. Our results demonstrate a conserved role for GRAS42/SCARECROW-LIKE 28 (SCL28)/DLT in BR signaling, clarify the role of this gene in GA signaling, and suggest mechanisms of tillering and leaf angle control by BR."
Authors: Kun Dong, Fuqing Wu, Siqi Cheng, Shuai Li, Feng Zhang, Xinxin Xing, Xin Jin, Sheng Luo, Miao Feng, Rong Miao, Yanqi Chang, Shuang Zhang, Xiaoman You, Peiran Wang, Xin Zhang, Cailin Lei, Yulong Ren, Shanshan Zhu, Xiuping Guo, Chuanyin Wu, Dong-Lei Yang, Qibing Lin, Zhijun Cheng and Jianmin Wan.
Molecular Plant (2024)
Abstract: "Although both protein arginine methylation (PRMT) and jasmonate (JA) signaling are crucial for regulating plant development, the relationship between these processes in spikelet development control remains unclear. Here, we utilized CRISPR/Cas9 technology to generate two OsPRMT6a loss-of-function mutants exhibiting various abnormal spikelet structures. Additionally, we found that OsPRMT6a could methylate arginine residues in the JA signal repressors OsJAZ1 and OsJAZ7. Arginine methylation of OsJAZ1 increased the affinity of OsJAZ1 for the JA receptors OsCOI1a and OsCOI1b in the presence of jasmonates (JAs), subsequently promoting the ubiquitination of OsJAZ1 by the SCFOsCOI1a/OsCOI1b complex and degradation via the 26S proteasome. This process ultimately released OsMYC2, a core transcriptional regulator in the JA signaling pathway, to activate or repress JA-responsive genes, thereby maintaining normal plant (spikelet) development. However, in the osprmt6a-1 mutant, reduced arginine methylation of OsJAZ1 impaired the interaction between OsJAZ1 and OsCOI1a/OsCOI1b in the presence of JAs. As a result, OsJAZ1 proteins became more stable, repressing JA responses, thus causing the formation of abnormal spikelet structures. Moreover, we discovered that JA signaling reduced the OsPRMT6a mRNA level in an OsMYC2-dependent manner, thereby establishing a negative feedback loop to balance JA signaling. Furthermore, we found that OsPRMT6a-mediated arginine methylation of OsJAZ1 likely serves as a switch to tune JA signaling to maintain normal spikelet development under harsh environmental conditions such as high temperatures. Thus, our study established a direct molecular link between arginine methylation and the JA signaling pathway.
Authors: Hideki Yoshida and Makoto Matsuoka.
Molecular Plant (2024)
Excerpts: "For example, brassinosteroids (BRs) positively affect plant growth and grain size but negatively regulate the grain number and plant density (Figure 1, top left; Deveshwar et al., 2020; Hwang et al., 2021). Therefore, a thorough understanding of the trade-off functions and regulatory mechanisms of phytohormones is a promising avenue for agricultural improvement."
"Zhang et al. (2024) revealed the molecular regulation of panicle branching by BRs through the cloning and functional analysis of a long-puzzling gene for the clustered spikelet (CL) phenotype. These observations demonstrated that the proper regulation of BR function resulted in higher rice yield."
"Zhang et al. (2024) successfully cloned a causal gene for CL using an elegant genomic analysis with both short- and long-read sequencing techniques. The CL trait is associated with complex structural variations in chromosomes that activate the expression of the BR catabolic gene BRASSINOSTEROID-DEFICIENT DWARF3 (BRD3) in secondary branch meristems."
"Zhang et al. (2024) also showed that introducing the genetic modification of the BRD3 promoter into modern rice varieties resulted in a higher rice yield with a high grain number due to CL and no other defects in field tests."
Authors: Kei Tsuzuki, Taiki Suzuki, Michio Kuruma, Kotaro Nishiyama, Ken-ichiro Hayashi, Shinya Hagihara and Yoshiya Seto.
bioRxiv (2024)
Abstract: "Root parasitic plants in the Orobancheceae, such as Striga and Orobanche, cause significant damage to crop production. The germination step of these root parasitic plants is induced by host-root-derived strigolactones (SLs). After germination, the radicles elongate toward the host and invade the host root. We have previously discovered that a simple amino acid, tryptophan (Trp), as well as its metabolite, the plant hormone indole-3-acetic acid (IAA), can inhibit radicle elongation of Orobanche minor. These results suggest that auxin plays a crucial role in the radicle elongation step in root parasitic plants. In this report, we used various auxin chemical probes to dissect the auxin function in the radicle growth of O. minor and Striga hermonthica. We found that synthetic auxins inhibited radicle elongation. In addition, auxin receptor antagonist, auxinole, rescued the inhibition of radicle growth by exogenous IAA. Moreover, a polar transport inhibitor of auxin, N-1-naphthylphthalamic acid (NPA), affected radicle tropism. We also proved that exogenously applied Trp is converted into IAA in O. minor seeds, and auxinole partly rescued this radicle elongation. Our data demonstrate a pivotal role of auxin in radicle growth. Thus, manipulation of auxin function in root parasitic plants should offer a useful approach to combat these parasites."
Authors: Tao Wang, Xuemin Ma, Ying Chen, Cuicui Wang, Zhenxiao Xia, Zixi Liu, Lihong Gao and Wenna Zhang.
Plant, Cell & Environment (2024)
Abstract: "Low temperature stress poses a significant challenge to the productivity of horticultural crops. The dynamic expression of cold-responsive genes plays a crucial role in plant cold tolerance. While NAC transcription factors have been extensively studied in plant growth and development, their involvement in regulating plant cold tolerance remains poorly understood. In this study, we focused on the identification and characterisation of SlNAC3 as the most rapid and robust responsive gene in tomato under low temperature conditions. Manipulating SlNAC3 through overexpression or silencing resulted in reduced or enhanced cold tolerance, respectively. Surprisingly, we discovered a negative correlation between the expression of CBF and cold tolerance in the SlNAC3 transgenic lines. These findings suggest that SlNAC3 regulates tomato cold tolerance likely through a CBF-independent pathway. Furthermore, we conducted additional investigations to identify the molecular mechanisms underlying SlNAC3-mediated cold tolerance in tomatoes. Our results revealed that SlNAC3 controls the transcription of ethylene biosynthetic genes, thereby bursting ethylene release in response to cold stress. Indeed, the silencing of these genes led to an augmentation in cold tolerance. This discovery provides valuable insights into the regulatory pathways involved in ethylene-mediated cold tolerance in tomatoes, offering potential strategies for developing innovative approaches to enhance cold stress resilience in this economically important crop species."
Authors: Wenli Hu, Rong Wang, Xiaohua Hao, Shaozhuang Li, Xinjie Zhao, Zijing Xie, Sha Wu, Liqun Huang, Ying Tan, Lianfu Tian and Dongping Li.
The Plant Journal (2024)
Significant Statement: The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight.
Abstract: "A fundamental question in developmental biology is how to regulate grain size to improve crop yields. Despite this, little is still known about the genetics and molecular mechanisms regulating grain size in crops. Here, we provide evidence that a putative protein kinase-like (OsLCD3) interacts with the S-adenosyl-L-methionine synthetase 1 (OsSAMS1) and determines the size and weight of grains. OsLCD3 mutation (lcd3) significantly increased grain size and weight by promoting cell expansion in spikelet hull, whereas its overexpression caused negative effects, suggesting that grain size was negatively regulated by OsLCD3. Importantly, lcd3 and OsSAMS1 overexpression (SAM1OE) led to large and heavy grains, with increased ethylene and decreased polyamines production. Based on genetic analyses, it appears that OsLCD3 and OsSAMS1 control rice grain size in part by ethylene/polyamine homeostasis. The results of this study provide a genetic and molecular understanding of how the OsLCD3-OsSAMS1 regulatory module regulates grain size, suggesting that ethylene/polyamine homeostasis is an appropriate target for improving grain size and weight."
Authors: Hongjiao Jiang, Lijun Xie, Zhiqun Gu, Hongyao Mei, Haohao Wang, Jing Zhang, Minmin Wang, Yiteng Xu, Chuanen Zhou and Lu Han.
The Plant Journal (2024)
Significance Statement: Auxins play important roles in various aspects of plant growth and development. In Medicago truncatula, orthologs of PIN1-mediated auxin transportation are involved in various developmental processes and amino acid biosynthesis and metabolism in seeds, which offers insights into the molecular mechanisms governing seed size regulation in crops.
Abstract: "The regulation of seed development is critical for determining crop yield. Auxins are vital phytohormones that play roles in various aspects of plant growth and development. However, its role in amino acid biosynthesis and metabolism in seeds is not fully understood. In this study, we identified a mutant with small seeds through forward genetic screening in Medicago truncatula. The mutated gene encodes MtPIN4, an ortholog of PIN1. Using molecular approaches and integrative omics analyses, we discovered that auxin and amino acid content significantly decreased in mtpin4 seeds, highlighting the role of MtPIN4-mediated auxin distribution in amino acid biosynthesis and metabolism. Furthermore, genetic analysis revealed that the three orthologs of PIN1 have specific and overlapping functions in various developmental processes in M. truncatula. Our findings emphasize the significance of MtPIN4 in seed development and offer insights into the molecular mechanisms governing the regulation of seed size in crops. This knowledge could be applied to enhance crop quality by targeted manipulation of seed protein regulatory pathways."
Authors: Guangwei Sun, Xuanhao Zhang, Yi Liu, Liguang Chai, Daisong Liu, Zhenguo Chen and Shiyou Lü.
Phyton (2024)
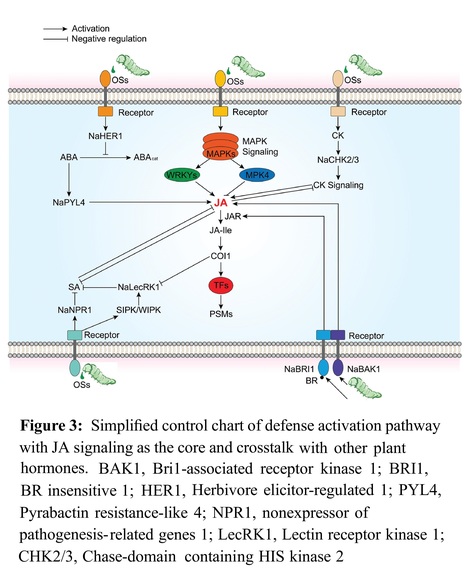
Abstract: "The Nicotiana genus, commonly known as tobacco, holds significant importance as a crucial economic crop. Confronted with an abundance of herbivorous insects that pose a substantial threat to yield, tobacco has developed a diverse and sophisticated array of mechanisms, establishing itself as a model of plant ecological defense. This review provides a concise overview of the current understanding of tobacco’s defense strategies against herbivores. Direct defenses, exemplified by its well-known tactic of secreting the alkaloid nicotine, serve as a potent toxin against a broad spectrum of herbivorous pests. Moreover, in response to herbivore attacks, tobacco enhances the discharge of volatile compounds, harnessing an indirect strategy that attracts the predators of the herbivores. The delicate balance between defense and growth leads to the initiation of most defense strategies only after a herbivore attack. Among plant hormones, notably jasmonic acid (JA), play central roles in coordinating these defense processes. JA signaling interacts with other plant hormone signaling pathways to facilitate the extensive transcriptional and metabolic adjustments in plants following herbivore assault. By shedding light on these ecological defense strategies, this review emphasizes not only tobacco’s remarkable adaptability in its natural habitat but also offers insights beneficial for enhancing the resilience of current crops."
Authors: Puja Ghosh and Aryadeep Roychoudhury.
Journal of Plant Growth Regulation (2024)
Abstract: "Plants are constantly exposed to a wide range of stress situations, such as biotic stressors caused by pathogenic microbes and herbivores, as well as abiotic stress factors like drought, salinity, and severe temperature. Plants have developed an intricate and sophisticated network of molecular signaling in response to various stresses that allow them to recognize and respond to challenging circumstances. Plant peptides have emerged as key participants in orchestrating biotic and abiotic stress responses, as well as taking part in signaling pathways for reactive oxygen species (ROS). ROS play a key role as secondary messengers in stress signaling and include chemicals, including superoxide, hydrogen peroxide, and singlet oxygen. This exhaustive review investigates the multiple functions of plant peptides in ROS signaling and stress responses and clarifies the interaction between plant peptide-mediated stress responses and ROS signaling. We have highlighted several plant peptide classes such as defense peptides, antimicrobial peptides and signaling peptides that contribute to the complex interaction between abiotic and biotic stress response pathways. A thorough overview of the complex regulatory network that permits plants to flourish under demanding environmental conditions is presented by synthesizing recent gains in our knowledge of the functions of plant peptides in stress responses and ROS signaling. We have also highlighted the fact that knowledge of such peptide-mediated signaling may be used for crop improvement that would increase agricultural sustainability and stress tolerance, ultimately boosting food security and environmental resilience on a global scale."
Authors: Akbar Ali, Krishan Kant, Navneet Kaur, Shalu Gupta, Parnika Jindal, Sarvajeet Singh Gill and M. Naeem.
South African Journal of Botany (2024)
Highlights: • Salicylic acid (SA) has encouraging effects on the growth and development of plants. • SA stimulates signalling in plants via several stress proteins and genes. • SA positively interacts with other phytohormones. • SA enhances plant tolerance towards abiotic stress.
Abstract: "Drastic change in global climatic conditions has significantly increased the frequency of abiotic stresses, such as different temperature regimes (high, low, or freezing stress), uneven precipitation leading to flooding or drought, soil salinization, cyclones and hurricanes which pose a major challenge to the crop productivity and food security. Therefore, it becomes imperative for the global science community to engineer stress tolerance in crop plants to ensure enough food for the globe. Plant growth regulators play an important function in stress management. One putative plant hormone that aids plants in coping with biotic and abiotic stressors is salicylic acid (SA). SA also cooperates with other phytohormones, such as gibberellins, auxins, abscisic acid, jasmonic acid, ethylene, polyamines, nitric oxide, and to counter the negative effects of environmental perturbations. Moreover, SA shields plants from oxidative stress by reducing the production of reactive oxygen species in challenging circumstances. Additionally, SA stimulates gas exchange, photosynthesis, and osmolyte synthesis in plants, which counteract the damage caused by ROS. Exogenous application of SA to agricultural crops including medicinal and aromatic plants improves their abiotic stress tolerance, either individually or in combination with other phytohormones. SA can stimulate the production of secondary metabolites by controlling the expression of stress-related genes, activating or regulating several key enzymes, and balancing the ion content. The present review summarizes the various mechanisms by which SA confers abiotic stress tolerance in plants through homeostasis, signalling, and crosstalk with other phytohormones."
Authors: Yoon Jeong Jang, Taehoon Kim, Makou Lin, Jeongim Kim, Kevin Begcy, Zhongchi Liu and Seonghee Lee.
bioRxiv (2024)
Abstract: "The plant hormone auxin plays a crucial role in regulating important functions in strawberry fruit development. Although a few studies have described the complex auxin biosynthetic and signaling pathway in wild diploid strawberry (Fragaria vesca), the molecular mechanisms underlying auxin biosynthesis and crosstalk in octoploid strawberry fruit development are not fully characterized. To address this knowledge gap, comprehensive transcriptomic analyses were conducted at different stages of fruit development and compared between the achene and receptacle to identify developmentally regulated auxin biosynthetic genes and transcription factors during the fruit ripening process. Similar to wild diploid strawberry, octoploid strawberry accumulates high levels of auxin in achene compared to receptacle. Consistently, genes functioning in auxin biosynthesis and conjugation, such as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAAs), YUCCA (YUCs), and GRETCHEN HAGEN 3 (GH3s) were found to be primarily expressed in the achene, with low expression in the receptacle. Interestingly, several genes involved in auxin transport and signaling like PIN-FORMED (PINs), AUXIN/INDOLE-3-ACETIC ACID proteins (Aux/IAAs), TRANSPORT INHIBITOR RESPONSE 1 / AUXIN-SIGNALING F-BOX (TIR/AFBs) and AUXIN RESPONSE FACTOR (ARFs) were more abundantly expressed in the receptacle. Moreover, by examining DEGs and their transcriptional profiles across all six developmental stages, we identified key auxin-related genes co-clustered with transcription factors from the NAM-ATAF1,2-CUC2/ WRKYGQK motif (NAC/WYKY), BASIC REGION/ LEUCINE ZIPPER motif (bZIP), and APETALA2/Ethylene Responsive Factor (AP2/ERF) groups. These results elucidate the complex regulatory network of auxin biosynthesis and its intricate crosstalk within the achene and receptacle, enriching our understanding of fruit development in octoploid strawberries."
Authors: Shammi Akter, Oscar Castaneda-Méndez and Jesús Beltrán
Current Opinion in Biotechnology (2024)
Highlights • Several challenges remain for the widespread use of synthetic biology in crops. • Synthetic gene expression circuitry is advancing in model plants. • Rapid enzyme discovery and testing are needed for plant metabolic engineering.
Abstract: "Plant synthetic biology (Plant SynBio) is an emerging field with the potential to enhance agriculture, human health, and sustainability. Integrating genetic tools and engineering principles, Plant SynBio aims to manipulate cellular functions and construct novel biochemical pathways to develop plants with new phenotypic traits, enhanced yield, and be able to produce natural products and pharmaceuticals. This review compiles research efforts in reprogramming plant developmental and biochemical pathways. We highlight studies leveraging new gene expression toolkits to alter plant architecture for improved performance in model and crop systems and to produce useful metabolites in plant tissues. Furthermore, we provide insights into the challenges and opportunities associated with the adoption of Plant SynBio in addressing complex issues impacting agriculture and human health."
|
Authors: Cong Li, Yanhang Chen, Qing Hu, Xiaolan Yang, Yunfeng Zhao, Yan Lin, Jianbo Yuan, Jinbao Gu, Yang Li, Jin He, Dong Wang, Bin Liu and Zhen-Yu Wang.
Plant Physiology (2024)
One-sentence summary: A pseudo-response regulator from the circadian system regulates a transcription factor involved in the abscisic acid-mediated drought stress response in soybean.
Abstract: "The circadian system plays a pivotal role in facilitating the ability of crop plants to respond and adapt to fluctuations in their immediate environment effectively. Despite the increasing comprehension of PSEUDO-RESPONSE REGULATORs (PRRs) and their involvement in the regulation of diverse biological processes, including circadian rhythms, photoperiodic control of flowering, and responses to abiotic stress, the transcriptional networks associated with these factors in soybean (Glycine max (L.) Merr.) remain incompletely characterized. In this study, we provide empirical evidence highlighting the significance of GmPRR3b as a crucial mediator in regulating the circadian clock, drought stress response, and abscisic acid (ABA) signaling pathway in soybeans. A comprehensive analysis of DNA affinity purification sequencing and transcriptome data identified 795 putative target genes directly regulated by GmPRR3b. Among them, a total of 570 exhibited a significant correlation with the response to drought, and eight genes were involved in both the biosynthesis and signaling pathways of ABA. Notably, GmPRR3b played a pivotal role in the negative regulation of the drought response in soybeans by suppressing the expression of abscisic acid responsive element-binding factor 3 (GmABF3). Additionally, the overexpression of GmABF3 exhibited an increased ability to tolerate drought conditions, and it also restored the hypersensitive phenotype of the GmPRR3b overexpressor. Consistently, studies on the manipulation of GmPRR3b gene expression and genome editing in plants revealed contrasting reactions to drought stress. The findings of our study collectively provide compelling evidence that emphasizes the significant contribution of the GmPRR3b-GmABF3 module in enhancing drought tolerance in soybean plants. Moreover, the transcriptional network of GmPRR3b provides valuable insights into the intricate interactions between this gene and the fundamental biological processes associated with plant adaptation to diverse environmental conditions."
Authors: Mahamud Hossain Al-Mamun, Christopher Ian Cazzonelli and Priti Krishna.
Frontiers in Plant Science (2024)
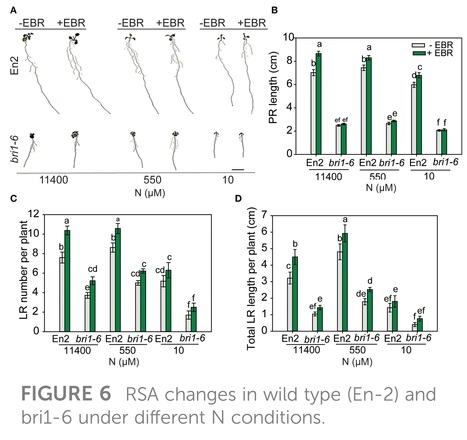
Abstract: "Plants modify their root system architecture (RSA) in response to nitrogen (N) deficiency. The plant steroidal hormone, brassinosteroid (BR), plays important roles in root growth and development. This study demonstrates that optimal levels of exogenous BR impact significant increases in lateral root length and numbers in Arabidopsis seedlings under mild N-deficient conditions as compared to untreated seedlings. The impact of BR on RSA was stronger under mild N deficiency than under N-sufficient conditions. The BR effects on RSA were mimicked in dominant mutants of BZR1 and BES1 (bzr1-1D and bes1-D) transcription factors, while the RSA was highly reduced in the BR-insensitive mutant bri1-6, confirming that BR signaling is essential for the development of RSA under both N-sufficient and N-deficient conditions. Exogenous BR and constitutive activity of BZR1 and BES1 in dominant mutants led to enhanced root meristem, meristematic cell number, and cortical cell length. Under mild N deficiency, bzr1-1D displayed higher fresh and dry shoot weights, chlorophyll content, and N levels in the shoot, as compared to the wild type. These results indicate that BR modulates RSA under both N-sufficient and N-deficient conditions via the transcription factors BES1/BZR1 module and confers tolerance to N deficiency."
Authors: Tomás Urzúa Lehuedé, Victoria Berdion Gabarain, Tomas Moyano, Lucia Ferrero, Miguel Angel Ibeas, Hernan Salinas-Grenet, Gerardo Núñez-Lillo, Romina Acha, Jorge Perez, Florencia Perotti, Virginia Natali Miguel, Fiorella Paola Spies, Miguel A. Rosas, Michitaro Shibata, Diana R. Rodríguez-García, Adrian A. Moreno, Keiko Sugimoto, Karen Sanguinet, Claudio Meneses, Raquel L. Chan, Federico Ariel, Jose M. Alvarez and José M. Estevez.
bioRxiv (2024)
Abstract: "The root hair (RH) cells can elongate to several hundred times their initial size, and are an ideal model system for investigating cell size control. Their development is influenced by both endogenous and external signals, which are combined to form a integrative response. Surprisingly, a low temperature condition of 10°C causes an increased RH growth in Arabidopsis and in several monocots, even when the development of the rest of the root and aerial parts of the plant are halted. Previously, we demonstrated a strong correlation between the growth response and a significant decrease in nutrient availability in the medium under low temperature conditions. However, the molecular basis responsible for receiving and transmitting signals related to the availability of nutrients in the soil, and their relation to plant development, remain largely unknown. We decided to further investigate the intricate molecular processes behind the particular responsiveness of this root cell type at low temperature. In this study, we have discovered a gene regulatory network (GRN) controlling early transcriptome responses to low temperature. This GNR is commanded by specific transcription factors (FTs), namely ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), a member of the homeodomain leucine zipper (HD-Zip I) group I 13 (AtHB13), the trihelix TF GT2-LIKE1 (GTL1), and a previously unidentified MYB-like TF (AT2G01060). Furthermore, we have identified four downstream RSL4 targets AtHB16, AtHB23, EARLY-RESPONSIVE TO DEHYDRATION 7 (ERD7) and ERD10 suggesting their participation in the regulation of RH development under these conditions. Functional analysis shows that such components of the RSL4-dependent transcriptional cascade influence the subsequent RH growth response to low temperature. These discoveries enhance our comprehension of how plants synchronize the RH growth in response to variations in temperature and nutrient availability at the cellular level."
Authors: Dongmei Yan and Huilan Yi.
Scientia Horticulturae (2024)
Highlights • Identifying key genes and pathways for postharvest preservation of grapes by RNA-seq. • SO2 prolongs the postharvest shelf life of table grapes by reducing cell wall degradation and enhancing defenses of the skins. • SO2-triggered secondary metabolism and defense responses in grape fruits contributed to postharvest preservation. • SO2-induced inhibition of ethylene signaling pathway helped to delay postharvest softening and aging of grape fruits.
Abstract: "Sulfur dioxide (SO2) is the most frequently used preservative for table grapes, yet the preservation mechanism remains unclear. To ascertain the specific genes and pathways involved in SO2 preservation, RNA-seq technology was employed to characterize the transcriptome profile of grapes during postharvest storage. A total of 22,288 genes were identified, of which 377 genes were differentially expressed (≥ 2-fold change) between SO2 and control groups, mainly enriched in secondary metabolism, plant-pathogen interactions, plant hormone signaling, etc. Numerous genes encoding pathogenesis-related (PR) proteins exhibited higher expression levels in SO2 group, while the activities of disease-resistant enzymes, such as β-1,3-glucanase (PR2) and chitinase (PR3) significantly increased by 28.9 % and 29.3 % (P < 0.05), indicating SO2-induced plant resistance. Exposure to SO2 also enhanced the expression of genes encoding essential enzymes of secondary metabolism, and significantly increased both the activities of critical enzymes for the biosynthesis of secondary metabolites, such as phenylalanine ammonia-lyase (PAL) and 4-coumarate-CoA ligase (4CL), and the contents of secondary metabolites such as total phenol, flavonoid, anthocyanin, and lignin, demonstrating an enhanced chemical and physical barriers in SO2-fumigated grapes. Meanwhile, the genes associated with ethylene signaling and cell wall degradation were down-regulated by SO2 preservative, and some ethylene-responsive elements were identified in the promoter regions of cell wall hydrolase genes, suggesting that SO2-inhibited ethylene signaling might contribute to maintaining cell wall integrity. Altogether, gene expression patterns and cellular physiological processes were altered in grapes exposed to SO2, which helped maintain fruit quality and prolong postharvest life by regulating the defense responses and fruit ageing."
Authors: Phanu T. Serivichyaswat, Abdul Kareem, Ming Feng and Charles W. Melnyk.
Plant Physiology (2024)
One-sentence summary: Auxin signaling in the cambial tissues promotes cell divisions and drives both tissue attachment and vascular differentiation during successful Arabidopsis grafting.
Abstract: "The strong ability of plants to regenerate wounds is exemplified by grafting when two plants are cut and joined together to grow as one. During graft healing, tissues attach, cells proliferate, and the vasculatures connect to form a graft union. The plant hormone auxin plays a central role, and auxin-related mutants perturb grafting success. Here, we investigated the role of individual cell types and their response to auxin during Arabidopsis (Arabidopsis thaliana) graft formation. By employing a cell-specific inducible misexpression system, we blocked auxin response in individual cell types using the bodenlos mutation. We found that auxin signaling in procambial tissues was critical for successful tissue attachment and vascular differentiation. In addition, we found that auxin signaling was required for cell divisions of the procambial cells during graft formation. Loss of function mutants in cambial pathways also perturbed attachment and phloem reconnection. We propose that cambial and procambial tissues drive tissue attachment and vascular differentiation during successful grafting. Our study thus refines our knowledge of graft development and furthers our understanding of the regenerative role of the cambium."
Author: Gwendolyn K. Kirschner.
The Plant Journal (2024)
Excerpts: "Udvardi and Sonali Roy, a former post-doctoral fellow with Udvardi at the Noble Research Institute and now an assistant professor at Tennessee State University, together with a team of experts on legume rhizobial symbiosis from Oklahoma, Cambridge and Shanghai, analysed the role of GLVs for nodule development and positioning in more detail (Roy et al., 2024)."
"Transcriptional reporter expression in transgenic hairy roots showed that the expression of the five GLVs overlapped at sites of nodule initiation (Figure 1b). Out of these, MtGLV6, MtGLV9 and MtGLV10 were induced by short-term nitrogen deprivation stress, suggesting a role in early stages of symbiosis, such as the dedifferentiation of cortical cells or the regulation of early nodulin genes."
"The nitrogen-induced GLV10 peptide could integrate the nitrogen signal from the soil to adaptation of the root architecture. GLV10p treatment also led to more microcolonies and infection thread initiations. The authors hypothesise that the increase in infection might be a compensation for the reduction in nodule number: The plant initiates more infection threads to make an optimal number of nodules and derive sufficient nitrogen."
Authors: Mengdi Fu, Xiaolei Yao, Xiaolin Li, Jing Liu, Mengyan Bai, Zijun Fang, Jiming Gong, Yuefeng Guan and Fang Xie.
The Plant Journal (2024)
Significance Statement: GmNLP1 and GmNLP4 activate nitrate-induced CLE peptides NIC1a/b to mediate nitrate-regulated root nodulation.
Abstract: "Symbiotic nitrogen fixation is an energy-intensive process, to maintain the balance between growth and nitrogen fixation, high concentrations of nitrate inhibit root nodulation. However, the precise mechanism underlying the nitrate inhibition of nodulation in soybean remains elusive. In this study, CRISPR-Cas9-mediated knockout of GmNLP1 and GmNLP4 unveiled a notable nitrate-tolerant nodulation phenotype. GmNLP1b and GmNLP4a play a significant role in the nitrate-triggered inhibition of nodulation, as the expression of nitrate-responsive genes was largely suppressed in Gmnlp1b and Gmnlp4a mutants. Furthermore, we demonstrated that GmNLP1b and GmNLP4a can bind to the promoters of GmNIC1a and GmNIC1b and activate their expression. Manipulations targeting GmNIC1a and GmNIC1b through knockdown or overexpression strategies resulted in either increased or decreased nodule number in response to nitrate. Additionally, transgenic roots that constitutively express GmNIC1a or GmNIC1b rely on both NARK and hydroxyproline O-arabinosyltransferase RDN1 to prevent the inhibitory effects imposed by nitrate on nodulation. In conclusion, this study highlights the crucial role of the GmNLP1/4-GmNIC1a/b module in mediating high nitrate-induced inhibition of nodulation."
Authors: Lu Liu, Yanzhi Liu, Xuehan Ji, Xia Zhao, Jun Liu and Ning Xu.
The Plant Journal (2024)
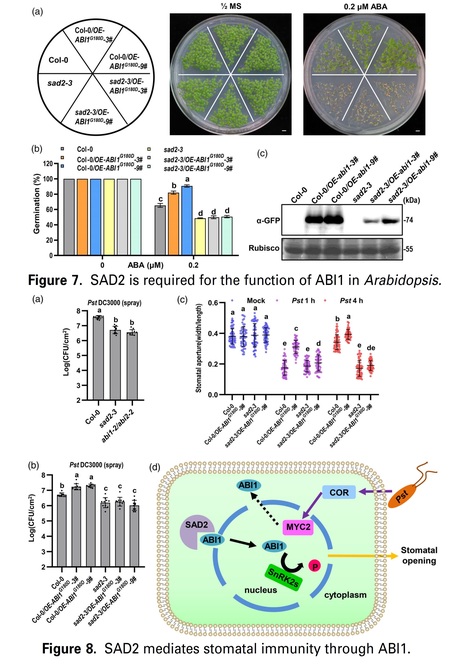
Significance Statement: Bacterial pathogens secret coronatine (COR) to recruit an importin β protein SAD2 for interfering ABA signaling in stomata and promote invasion.
Abstract: "Stomatal immunity plays an important role during bacterial pathogen invasion. Abscisic acid (ABA) induces plants to close their stomata and halt pathogen invasion, but many bacterial pathogens secrete phytotoxin coronatine (COR) to antagonize ABA signaling and reopen the stomata to promote infection at early stage of invasion. However, the underlining mechanism is not clear. SAD2 is an importin β family protein, and the sad2 mutant shows hypersensitivity to ABA. We discovered ABI1, which negatively regulated ABA signaling and reduced plant sensitivity to ABA, was accumulated in the plant nucleus after COR treatment. This event required SAD2 to import ABI1 to the plant nucleus. Abolition of SAD2 undermined ABI1 accumulation. Our study answers the long-standing question of how bacterial COR antagonizes ABA signaling and reopens plant stomata during pathogen invasion."
Authors: Lei Liang and Xiangyang Hu.
Phyton (2024)
Abstract: "With the rapid development of modern molecular biology and bioinformatics, many studies have proved that transcription factors play an important role in regulating the growth and development of plants. SPATULA (SPT) belongs to the bHLH transcription family and participates in many processes of regulating plant growth and development. This review systemically summarizes the multiple roles of SPT in plant growth, development, and stress response, including seed germination, flowering, leaf size, carpel development, and root elongation, which is helpful for us to better understand the functions of SPT."
Authors: Rina Saito, Yuho Nishizato, Tsumugi Kitajima, Misuzu Nakayama, Yousuke Takaoka, Nobuki Kato and Minoru Ueda.
bioRxiv (2024)
Abstract: "(+)-cis-12-oxo-phytodienoic acid (cis-OPDA) is a biosynthetic precursor of the plant hormone (+)-7-iso-jasmonoyl-L-isoleucine (JA-Ile). It functions as an endogenous chemical signal independent of the JA-Ile receptor COI1-JAZ in Arabidopsis thaliana. The bioactive form of cis-OPDA that induces COI1-JAZ-independent gene expression remains unknown. In this study, we hypothesized that the genuine bioactive forms of cis-OPDA are the downstream metabolites, which upregulate the expression of the OPDA marker genes such as ZAT10/ERF5 in a JA-Ile-independent manner. These downstream metabolites function independently of the JA-Ile-COI1-JAZ-MYCs canonical jasmonate signaling module, and its electrophilic nature is essential for its bioactivity."
Authors: Baris Uzilday, Kaori Takahashi, Akie Kobayashi, Rengin Ozgur Uzilday, Nobuharu Fujii, Hideyuki Takahashi and Ismail Turkan.
Plants (2024)
Abstract: "Plant roots exert hydrotropism in response to moisture gradients to avoid drought stress. The regulatory mechanism underlying hydrotropism involves novel regulators such as MIZ1 and GNOM/MIZ2 as well as abscisic acid (ABA), reactive oxygen species (ROS), and Ca2+ signaling. ABA, ROS, and Ca2+ signaling are also involved in plant responses to drought stress. Although the mechanism of moisture gradient perception remains largely unknown, the sensory apparatus has been reported to reside in the root elongation zone rather than in the root cap. In Arabidopsis roots, hydrotropism is mediated by the action of MIZ1 and ABA in the cortex of the elongation zone, the accumulation of ROS at the root curvature, and the variation in the cytosolic Ca2+ concentration in the entire root tip including the root cap and stele of the elongation zone. Moreover, root exposure to moisture gradients has been proposed to cause asymmetric ABA distribution or Ca2+ signaling, leading to the induction of the hydrotropic response. A comprehensive and detailed analysis of hydrotropism regulators and their signaling network in relation to the tissues required for their function is apparently crucial for understanding the mechanisms unique to root hydrotropism. Here, referring to studies on plant responses to drought stress, we summarize the recent findings relating to the role of ABA, ROS, and Ca2+ signaling in hydrotropism, discuss their functional sites and plausible networks, and raise some questions that need to be answered in future studies."
Authors: Yuanyuan Wu, Ying Sun, Wanmin Wang, Zizhao Xie, Chenghang Zhan, Liang Jin and Junli Huang.
Plant Physiology and Biochemistry (2024)
Highlights: • OsJAZ10 negatively modulates rice drought stress tolerance. • OsJAZ10 physically interacts with OsMYC2, and inhibits OsMYC2 transcriptional activation on ABA- and JA-biosynthetic genes, thus repressing ABA and JA response. • OsJAZ10 inhibits the transmission of osmotic stress-elicited systematic electrical signals, which is in parallel to the hormone response.
Abstract: "Jasmonic acid (JA) plays crucial functions in plant stress response, and the synergistic interaction between JA and abscisic acid (ABA) signaling is implicated to help plants adapt to environmental challenges, whereas the underlying molecular mechanism still needs to be revealed. Here, we report that OsJAZ10, a repressor in the JA signaling, represses rice drought tolerance via inhibition of JA and ABA biosynthesis. Function loss of OsJAZ10 markedly enhances, while overexpression of OsJAZ10ΔJas reduces rice drought tolerance. The osjaz10 mutant is more sensitive to exogenous ABA and MeJA, and produces higher levels of ABA and JA after drought treatment, indicating OsJAZ10 represses the biosynthesis of these two hormones. Mechanistic study demonstrated that OsJAZ10 physically interacts with OsMYC2. Transient transcriptional regulation assays showed that OsMYC2 activates the expression of ABA-biosynthetic gene OsNCED2, JA-biosynthetic gene OsAOC, and drought-responsive genes OsRAB21 and OsLEA3, while OsJAZ10 prevents OsMYC2 transactivation of these genes. Further, electrophoretic mobility shift assay (EMSA) confirmed that OsMYC2 directly binds to the promoters of OsNCED2 and OsRAB21. Electrical activity has been proposed to activate JA biosynthesis. Interestingly, OsJAZ10 inhibits the propagation of osmotic stress-elicited systemic electrical signals, indicated by the significantly increased PEG-elicited slow wave potentials (SWPs) in osjaz10 mutant, which is in accordance with the elevated JA levels. Collectively, our findings establish that OsJAZ10 functions as a negative regulator in rice drought tolerance by repressing JA and ABA biosynthesis, and reveal an important mechanism that plant integrates electrical events with hormone signaling to enhance the adaption to environmental stress."
|


 Your new post is loading...
Your new post is loading...