Your new post is loading...

|
Scooped by
Juan Lama
|
The Omicron subvariant BA.2 accounts for a large majority of the SARS-CoV-2 infection worldwide today. However, its recent descendants BA.2.12.1 and BA.4/5 have surged dramatically to become dominant in the United States and South Africa, respectively. That these novel Omicron subvariants carry additional mutations in their spike proteins raises concerns that they may further evade neutralizing antibodies, thereby further compromising the efficacy of our COVID-19 vaccines and therapeutic monoclonals. We now report findings from a systematic antigenic analysis of these surging Omicron subvariants. BA.2.12.1 is only modestly (1.8-fold) more resistant to sera from vaccinated and boosted individuals than BA.2. On the other hand, BA.4/5 is substantially (4.2-fold) more resistant and thus more likely to lead to vaccine breakthrough infections. Mutation at spike residue L452 found in both BA.2.12.1 and BA.4/5 facilitates escape from some antibodies directed to the so-called Class 2 and Class 3 regions of the receptor-binding domain (RBD). The F486V mutation found in BA.4/5 facilitates escape from certain Class 1 and Class 2 antibodies to the RBD but compromises the spike affinity for the cellular receptor ACE2. The R493Q reversion mutation, however, restores receptor affinity and consequently the fitness of BA.4/5. Among therapeutic antibodies authorized for clinical use, only bebtelovimab (LY-COV1404) retains full potency against both BA.2.12.1 and BA.4/5. The Omicron lineage of SARS-CoV-2 continues to evolve, successively yielding subvariants that are not only more transmissible but also more evasive to antibodies. Preprint in bioRxiv (May 26, 2022): https://doi.org/10.1101/2022.05.26.493517

|
Scooped by
Juan Lama
|
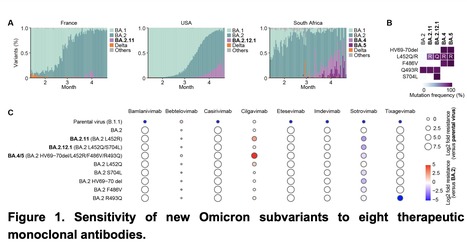
As of May 2022, Omicron BA.2 variant is the most dominant variant in the world. Thereafter, Omicron subvariants have emerged and some of them began outcompeting BA.2 in multiple countries. For instance, Omicron BA.2.11, BA.2.12.1 and BA.4/5 subvariants are becoming dominant in France, the USA and South Africa, respectively. In this study, we evaluated the sensitivity of these new Omicron subvariants (BA.2.11, BA.2.12.1 and BA.4/5) to eight therapeutic monoclonal antibodies (bamlanivimab, bebtelovimab, casirivimab, cilgavimab, etesevimab, imdevimab, sotrovimab and tixagevimab). Notably, we showed that although cilgavimab is antiviral against BA.2, BA.4/5 exhibits higher resistance to this antibody compared to BA.2. Since mutations are accumulated in the spike proteins of newly emerging SARS-CoV-2 variants, we suggest the importance of rapid evaluation of the efficiency of therapeutic monoclonal antibodies against novel SARS-CoV-2 variants. Preprint available at bioRxiv (May 3, 2022): https://doi.org/10.1101/2022.05.03.490409

|
Scooped by
Juan Lama
|
SARS-CoV-2, the virus that causes COVID-19, is constantly changing and accumulating mutations in its genetic code over time. New variants of SARS-CoV-2 are expected to continue to emerge. Some variants will emerge and disappear, while others will emerge and continue to spread and may replace previous variants. To identify and track SARS-CoV-2 variants, CDC uses genomic surveillance. CDC’s national genomic surveillance system collects SARS-CoV-2 specimens for sequencing through the National SARS-CoV-2 Strain Surveillance (NS3) program, as well as SARS-CoV-2 sequences generated by commercial or academic laboratories contracted by CDC and state or local public health laboratories. Virus genetic sequences are analyzed and classified as a particular variant. The proportion of variants in a population are calculated nationally, by HHS region, and by jurisdiction. The thousands of sequences analyzed every week through CDC’s national genomic sequencing and bioinformatics efforts fuel the comprehensive and population-based U.S. surveillance system established to identify and monitor the spread of variants. Rapid virus genomic sequencing data combined with phenotypic data are further used to determine whether COVID-19 tests, treatments, and vaccines authorized or approved for use in the United States will work against emerging variants.
|

|
Scooped by
Juan Lama
|
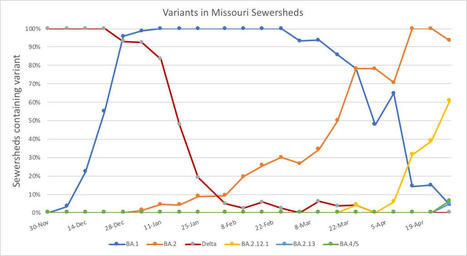
As cases rise in the U.S. and other parts of the world, Omicron subvariants are a continued culprit. Experts are watching closely to see if further mutations of the virus may become even more contagious, or may gain the ability to evade immunity from prior infections and vaccinations. BA.4 and BA.5 in South Africa When South African scientists sounded the alarm about Omicron in November, they identified three versions of the variant, called BA.1, BA.2, and BA.3. BA.1 first spread rapidly through South Africa and the rest of the world in December and January; BA.2 then outcompeted BA.1 to become the dominant strain globally. Now, South African scientists are sounding the alarm again: this time about new Omicron lineages, dubbed BA.4 and BA.5. These two lineages have driven another new surge in the country, with both cases and hospital admissions rising sharply in recent weeks. The surge might be starting to slow, as of late this week, but it’s unclear if this trend will continue. South Africa saw a huge BA.1 surge in November and December, then didn’t see much of a BA.2 bump—likely because so many people infected with BA.1 had immunity to this variant. But BA.4 and BA.5 may be a different story, according to a preprint from Alex Sigal and colleagues at the Africa Health Research Institute. Sigal and his collaborators tested neutralizing antibodies—a commonly-studied aspect of immune system protection—from BA.1 against BA.4 and BA.5. They found that a BA.1 infection offered relatively limited protection against BA.4 and BA.5, especially if the person who had BA.1 was unvaccinated. “BA.4 and BA.5 have potential to result in a new infection wave,” the authors wrote. This study is a preprint, not yet peer-reviewed. But it’s still a major warning sign for the U.S. and other countries: watch out for BA.4 and BA.5. BA.2 subvariants in the U.S. Meanwhile, here in the U.S., BA.2 continues to mutate and spread rapidly. The BA.2.12.1 subvariant, first identified by the New York State health department in mid-April as a sublineage that spreads even faster than BA.2, is now causing more than one-third of new COVID-19 cases in the country, according to CDC estimates. In New York and New Jersey, BA.2.12.1 is causing over 60% of new cases; it’s no coincidence that these states are also reporting some of the highest case and hospitalization rates in the country right now. New England, mid-Atlantic, South, and Midwest states are also seeing high proportions of BA.2.12.1. The CDC’s variant proportions estimates don’t yet include BA.4 and BA.5, but other reports suggest that these subvariants are already in the U.S. and starting to compete with our homegrown BA.2 lineages. Marc Johnson, a leading wastewater expert in Missouri, posted on Twitter yesterday that he’s seeing “a circus of Omicron sublineages” in his state, including BA.4 and BA.5. Also worth noting: a new U.S. study (shared as a preprint last week) found that, actually, Omicron is not inherently less severe than other variants. In comparing hospitalization and mortality risks after accounting for vaccination and medical risk factors, the researchers behind this study found little difference between the Omicron wave and prior periods. While this study also has yet to be peer-reviewed, it doesn’t bode well for future Omicron-driven surges.

|
Scooped by
Juan Lama
|
Recent emergence of SARS-CoV-2 Omicron sublineages BA.2.12.1, BA.2.13, BA.4 and BA.5 all contain L452 mutations and show potential higher transmissibility over BA.2. The new variants' receptor binding and immune evasion capability require immediate investigation, especially on the role of L452 substitutions. Herein, coupled with structural comparisons, we showed that BA.2 sublineages, including BA.2.12.1 and BA.2.13, exhibit increased ACE2-binding affinities compared to BA.1; while BA.4/BA.5 shows the weakest receptor-binding activity due to F486V and R493Q reversion. Importantly, compared to BA.2, BA.2.12.1 and BA.4/BA.5 exhibit stronger neutralization escape from the plasma of 3-dose vaccinees and, most strikingly, from vaccinated BA.1 convalescents. To delineate the underlying evasion mechanism, we determined the escaping mutation profiles, epitope distribution and Omicron sublineage neutralization efficacy of 1640 RBD-directed neutralizing antibodies (NAbs), including 614 isolated from BA.1 convalescents. Interestingly, post-vaccination BA.1 infection mainly recalls wildtype (WT) induced humoral memory and elicits antibodies that neutralize both WT and BA.1. These cross-reactive NAbs are significantly enriched on non-ACE2-competing epitopes; and surprisingly, the majority are undermined by R346 and L452 substitutions, namely R346K (BA.1.1), L452M (BA.2.13), L452Q (BA.2.12.1) and L452R (BA.4/BA.5), suggesting that R346K and L452 mutations appeared under the immune pressure of Omicron convalescents. Nevertheless, BA.1 infection can also induce new clones of BA.1-specific antibodies that potently neutralize BA.1 but do not respond to WT SARS-CoV-2, due to the high susceptibility to N501, N440, K417 and E484. However, these NAbs are largely escaped by BA.2 sublineages and BA.4/BA.5 due to D405N and F486V, exhibiting poor neutralization breadths. As for therapeutic NAbs, LY-CoV1404 (Bebtelovimab) and COV2-2130 (Cilgavimab) can still effectively neutralize BA.2.12.1 and BA.4/BA.5, while the S371F, D405N and R408S mutations carried by BA.2/BA.4/BA.5 sublineages would undermine most broad sarbecovirus NAbs. Together, our results indicate that Omicron can evolve mutations to specifically evade humoral immunity elicited by BA.1 infection. The continuous evolution of Omicron poses great challenges to SARS-CoV-2 herd immunity and suggests that BA.1-derived vaccine boosters may not be ideal for achieving broad-spectrum protection. Preprint available in bioRxiv (May 02, 2022): https://doi.org/10.1101/2022.04.30.489997
|



 Your new post is loading...
Your new post is loading...