Your new post is loading...

|
Scooped by
Juan Lama
|
The SARS-CoV-2 variant JN.1 swiftly became the global dominant strain due to a spike protein Leu455Ser substitution, boosting transmissibility and immune-escape capabilities, surpassing its predecessor BA.2.86 and other variants. These alterations have resulted in a surge of COVID-19 cases, reflected in wastewater-surveillance data surpassing rates, observed during the initial omicron wave. However, concerns persist that JN.1 might have an increased capacity to replicate in the gut, potentially leading to infected individuals shedding a higher number of viral copies than previously seen. As there is currently a lack of available data for fecal viral shedding, we are presenting the initial longitudinal and quantitative faecal shedding data for SARS-CoV-2 RNA in individuals infected with XBB.1.5, EG.5.1, HV.1, JD.1.1, BA.2.86, and JN.1. 856 faecal samples were obtained from 113 non-hospitalised individuals with confirmed PCR positivity for SARS-CoV-2 RNA. Variants were identified through Sanger sequencing. Detailed protocols for processing and extracting SARS-CoV-2 RNA from stool samples are provided in the appendix... Published in The Lancet Infectious Diseases (March 21, 2024):

|
Scooped by
Juan Lama
|
The SARS-CoV-2 BA.2.86 lineage, first identified in August 2023, is phylogenetically distinct from the currently circulating SARS-CoV-2 Omicron XBB lineages, including EG.5.1 and HK.3. Comparing to XBB and BA.2, BA.2.86 carries more than 30 mutations in the spike (S) protein, indicating a high potential for immune evasion. BA.2.86 has evolved and its descendant, JN.1 (BA.2.86.1.1), emerged in late 2023. JN.1 harbors S:L455S and three mutations in non-S proteins. S:L455S is a hallmark mutation of JN.1: we have recently shown that HK.3 and other "FLip" variants carry S:L455F, which contributes to increased transmissibility and immune escape ability compared to the parental EG.5.1 variant. Here, we investigated the virological properties of JN.1. Preprint in bioRxiv (Dec. 9, 2023): https://doi.org/10.1101/2023.12.08.570782

|
Scooped by
Juan Lama
|
Background: Subvariants of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) omicron XBB-lineage have the potential to escape immunity provided by prior infection or vaccination. For Covid-19 immunizations beginning in the Fall 2023, the U.S. FDA has recommended updating to a monovalent omicron XBB.1.5-containing vaccine. Methods: In this ongoing, phase 2/3 study participants were randomized 1:1 to receive 50-µg doses of mRNA-1273.815 monovalent (50-µg omicron XBB.1.5 spike mRNA) or mRNA-1273.231 bivalent (25-µg omicron XBB.1.5 and 25-µg omicron BA.4/BA.5 spike mRNAs) vaccines, administered as 5th doses, to adults who previously received a primary series and 3rd dose of an original mRNA coronavirus disease 2019 (Covid-19) vaccine, and a 4th dose of a bivalent (omicron BA.4/BA.5 and original SARS-CoV-2) vaccine. Interim safety and immunogenicity data 15 days post-vaccination are presented. Results: In April 2023, participants received mRNA-1273.815 (n=50) and mRNA-1273.231 (n=51). The median intervals from the prior dose of BA.4/BA.5-containing bivalent vaccine were 8.2 and 8.3 months for the mRNA-1273.815 and mRNA-1273.231 groups, respectively. Both vaccines increased neutralizing antibody (nAb) geometric mean titers against all variants tested at day 15 post-booster nAb compared to pre-booster levels. Geometric mean fold-rises from pre-booster titers after the monovalent booster were numerically higher against XBB.1.5, XBB.1.16 and SARS-CoV-2 (D614G) than those of the bivalent booster and were comparable against BA.4/BA.5 and BQ1.1 variants for both vaccines. The monovalent vaccine also elicited nAb responses against omicron XBB.2.3.2, EG.5.1, FL.1.5.1 and BA.2.86 that were similar to those against XBB.1.5 in a subset (n=20) of participants. The occurrence of solicited adverse reactions and unsolicited adverse events were overall similar to those previously reported for the original mRNA-1273 50-µg and omicron BA.4/BA.5-containing bivalent mRNA-1273 vaccines. Conclusion: In this interim analysis, XBB.1.5-containing monovalent and bivalent vaccines elicited potent neutralizing responses against variants of the omicron XBB-lineage (XBB.1.5, XBB.1.6, XBB.2.3.2, EG.5.1, and FL.1.5.1) as well as the recently emerged BA.2.86 variant. The safety profile of the XBB.1.5-containing vaccine was consistent with those of prior vaccines. These results overall indicate that the XBB.1.5-containing mRNA-1273.815 vaccine has the potential to provide protection against these emerging variants and support the Covid-19 vaccine update in 2023-2024 to a monovalent XBB.1.5-containing vaccine. Preprint avilable in medRxiv (Sept. 7, 2023): https://doi.org/10.1101/2023.08.22.23293434

|
Scooped by
Juan Lama
|
Research is under way to determine whether the mutation-laden lineage BA.2.86 is nothing to worry about — or has the potential to spread globally. Researchers are racing to determine whether a highly mutated coronavirus variant that has popped up in three continents will be a global concern — or much ado about nothing. Several laboratories detected the variant last week, and it has been named BA.2.86. Although the lineage seems to be exceedingly rare, it is very different from other circulating variants and carries numerous changes to its spike protein, a key focus of the body’s immune attack on the SARS-CoV-2 virus. To many scientists, the emergence of BA.2.86 is reminiscent of the early days of the Omicron variant in late 2021, when scientists in southern Africa noticed a weird-looking lineage that quickly went global. “There’s a little bit of déjà vu all over again,” says Adam Lauring, a virologist and infectious-disease physician at the University of Michigan in Ann Arbor, whose lab identified one person infected with BA.2.86. Successive COVID-19 waves and booster vaccine roll-outs mean global immunity to SARS-CoV-2 is higher and broader than ever, and most scientists do not expect BA.2.86 to have the same impact as Omicron’s arrival. “There’s good reason to think it won’t be like the Omicron wave, but it’s early days,” Lauring adds. What do we know about BA.2.86? As of 21 August, the variant had been linked to 6 cases in 4 countries: Israel, Denmark, the United Kingdom and the United States. The World Health Organization in Geneva, Switzerland, has designated BA.2.86 as a variant under monitoring. “Almost certainly there are going to be other cases that will start popping up,” adds Lauring. The lineage seems to be descended from an Omicron subvariant called BA.2, which caused large case spikes in early 2022. However, the BA.2.86 spike protein — the molecule that SARS-CoV-2 uses to enter cells — carries 34 changes relative to BA.2. Large numbers of spike mutations have been observed in people with long-term SARS-CoV-2 infections, and it is likely that BA.2.86 also emerged from one such chronic infection, says Jesse Bloom, a viral evolutionary biologist at the Fred Hutchinson Cancer Center in Seattle, Washington. Why are scientists following BA.2.86 so closely? After Omicron appeared, SARS-CoV-2 evolution began to follow a somewhat predictable course: successful new variants emerged from circulating lineages after gaining a few key mutations that enabled their spread. BA.2.86, by contrast, is drastically different from other widespread coronavirus variants, reminiscent of Omicron and early pandemic variants including Alpha and Delta. “Just like Omicron was a little out of left field, this BA.2.86 is little out of left field,” says Ashish Jha, a public-health researcher at Brown University in Providence, Rhode Island, and the former White House COVID-19 Response Coordinator. “There is enough here to get us all to start paying attention.” Many of BA.2.86’s changes are in regions of the spike protein targeted by the body’s potent infection-blocking, or neutralizing, antibodies, says Bloom, who posted a preliminary analysis of the variant last week. For this reason, there is a good chance that the variant will be able to escape some of the neutralizing antibodies triggered by previous infections and vaccine boosters. Another feature of BA.2.86 that has scientists paying attention is its geographical distribution. None of the cases seem to be linked — including three infections in Denmark that were detected in different parts of the country. This suggests that variant may already be fairly widespread, says Bloom. “It’s got to have been transmitting a fair amount.” The UK Health Security Agency said that there is a case in a person with no recent travel history, “suggesting a degree of community transmission within the UK”. What do researchers want to find out? Labs worldwide are now scouring patient samples as well as wastewater to get a sense of how widespread BA.2.86 is. “We want to try and understand how much of this lineage is out there,” says Lauring. If the current trickle of new confirmed cases turns into a flood, it will be a sign that the variant has the potential to compete with other circulating SARS-CoV-2 variants, including a more common lineage called EG.5, and cause a global spike in infections. Virology labs in Denmark and the United Kingdom say they are trying to isolate BA.2.86 from patient samples. Such work — and studies with safe models of SARS-CoV-2 called pseudoviruses — will help researchers to gauge the variant’s ability to evade neutralizing antibodies triggered by prior infections and vaccines. Bloom says that he is especially interested in seeing the extent to which BA.2.86 can evade neutralizing antibodies triggered by a recent infection with a variant called XBB.1.5, because the latest COVID-19 booster vaccines are based on that variant’s spike sequence. “If something like [BA.2.86] became widespread. I think you’d want to start thinking about updating the vaccine,” says Bloom. Should the public be concerned about BA.2.86? “I don’t think anybody needs to be alarmed by this,” stresses Bloom. “The most likely scenario is that this variant fizzles out, and in a month, nobody other than people like me even remember that it existed.” Even if BA.2.86 becomes widespread and proves adept at dodging neutralizing antibodies — which looks likely based on its set of spike mutations — other forms of immunity are likely to keep most people from getting seriously ill if they are infected, Bloom adds. Jha says scientists should pay close attention to the variant. But he thinks the chances are “exceedingly low” that it will turn out to be any more severe than existing variants or cause the level of disruption seen with the first Omicron waves, owing to widespread immunity. The appearance of BA.2.86 has caught scientists by surprise — and shows that SARS-CoV-2 still has more tricks up its sleeve that researchers will want to understand. “We’ll see if it’s important beyond that in terms of its public health impact,” says Lauring. doi: https://doi.org/10.1038/d41586-023-02656-9
|

|
Scooped by
Juan Lama
|
tThe SARS-CoV-2 saltation variant BA.2.86, which was quickly designated as a variant under monitoring after its emergence, has garnered global attention. Although BA.2.86 did not show substantial humoral immune escape and growth advantage compared with current dominant variants, such as EG.5.1 and HK.3, it showed remarkably high ACE2 binding affinity. This increased binding affinity, coupled with its distinct antigenicity, could enable BA.2.86 to accumulate immune-evasive mutations during low-level populational transmission, akin to the previous evolution from BA.2.75 to CH.1.1 and XBB. With just one additional receptor binding domain mutation (L455S) compared to its predecessor BA.2.86, the JN.1 variant rapidly became predominant in France (figure A; appendix 1 p 12), surpassing both BA.2.86 and the so-called FLip (L455F+F456L) strains. A thorough investigation into the immune evasion capability of JN.1, particularly given its few additional mutations, is imperative. Published in The Lancet Infectious Diseases (Dec. 15, 2023): https://doi.org/10.1016/S1473-3099(23)00744-2

|
Scooped by
Juan Lama
|
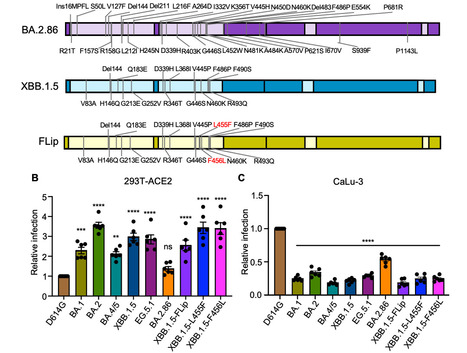
Evolution of SARS-CoV-2 requires the reassessment of current vaccine measures. Here, we characterized BA.2.86 and the XBB-lineage variant FLip by investigating their neutralization alongside D614G, BA.1, BA.2, BA.4/5, XBB.1.5, and EG.5.1 by sera from 3-dose vaccinated and bivalent vaccinated healthcare workers, XBB.1.5-wave infected first responders, and monoclonal antibody (mAb) S309. We assessed the biology of the variant Spikes by measuring viral infectivity and membrane fusogenicity. BA.2.86 is less immune evasive compared to FLip and other XBB variants, consistent with antigenic distances. Importantly, distinct from XBB variants, mAb S309 was unable to neutralize BA.2.86, likely due to a D339H mutation based on modeling. BA.2.86 had relatively high fusogenicity and infectivity in CaLu-3 cells but low fusion and infectivity in 293T-ACE2 cells compared to some XBB variants, suggesting a potentially differences conformational stability of BA.2.86 Spike. Overall, our study underscores the importance of SARS-CoV-2 variant surveillance and the need for updated COVID-19 vaccines. Preprint available in bioRxiv (Sept. 12, 2023): https://doi.org/10.1101/2023.09.11.557206

|
Scooped by
Juan Lama
|
BA.2.86, nicknamed Pirola, causing concern among scientists because of fear it could be more transmissible. The latest Covid-19 variant, BA.2.86, appears to be spreading in the UK, health surveillance data suggests. The variant, nicknamed Pirola, has prompted concern among scientists because of the high number of mutations it carries, which raises the possibility that it could evade the immune system more easily or be more transmissible. In a briefing note on Friday, the UK Health Security Agency (UKHSA) said that an outbreak at a care home in Norfolk and other cases across the UK indicated there was likely to be community transmission of the strain, but said it was too early to judge the full extent of its spread. In an outbreak of Covid-19 in a care home in Norfolk at the end of August, 33 out of 38 residents tested positive for the virus, along with 12 members of staff, the UKHSA said. One resident needed hospital treatment but no deaths were reported. Laboratory tests later showed that 22 residents had the BA.2.86 variant, along with six staff. The outbreak was “an early indicator” that the variant may be sufficiently transmissible to have an effect in close-contact settings, the UKHSA said, though it was too early to draw any conclusions about how BA.2.86 would behave in the wider UK population. Twenty-nine of the 33 residents at the care home who tested positive for Covid-19 have recovered, along with all members of staff, the UKHSA added. Dr Renu Bindra, the UKHSA incident director, said that while BA.2.86 had a “significant number of mutations” compared with other variants circulating, the data so far was “too limited to draw firm conclusions” about the impact this would have on the transmissibility or severity of the virus. “It is clear that there is some degree of widespread community transmission, both in the UK and globally, and we are working to ascertain the full extent of this,” she said. “In the meantime, it remains vital that all those eligible come forward to receive their autumn vaccine as soon as it is offered to them.” Some early lab-based evidence has eased initial concerns about the potential of BA.2.86 to cause a new global wave of infection, as happened with the emergence of Omicron. A pre-print study, from researchers in China, found that BA.2.86 is not as efficient at infecting cells in the lab compared with other circulating Omicron subvariants. Another pre-print study from researchers in Sweden found only modest drops in how well serum from blood donors could neutralise BA.2.86 compared with other variants. The latest Covid-19 vaccine booster programme has been brought forward from October to September as a precaution against BA.2.86. The booster programme will begin in England on 11 September, with jabs offered first to residents of adult care homes and clinically vulnerable people, before it is extended to everyone in the UK aged 65 and above. See also latest UK SARS-CoV-2 Surveillance Report (Sept. 4, 2023): https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings/sars-cov-2-variant-surveillance-and-assessment-technical-briefing-53
|
 Your new post is loading...
Your new post is loading...