Cryo-electron microscopy by University of Alabama at Birmingham researchers has exposed the structure of a bacterial virus with unprecedented detail. This is the first structure of a virus able to infect Staphylococcus epidermidis, and high-resolution knowledge of structure is a key link between viral biology and potential therapeutic use of the virus to quell bacterial infections. Bacteriophages or "phages" is the terms used for viruses that infect bacteria. The UAB researchers, led by Terje Dokland, Ph.D., in collaboration with Asma Hatoum-Aslan, Ph.D., at the University of Illinois Urbana-Champaign, have described atomic models for all or part of 11 different structural proteins in phage Andhra. The study is published in Science Advances. Andhra is a member of the picovirus group. Its host range is limited to S. epidermidis. This skin bacterium is mostly benign but also is a leading cause of infections of indwelling medical devices. "Picoviruses are rarely found in phage collections and remain understudied and underused for therapeutic applications," said Hatoum-Aslan, a phage biologist at the University of Illinois. With emergence of antibiotic resistance in S. epidermidis and the related pathogen Staphylococcus aureus, researchers have renewed interest in potentially using bacteriophages to treat bacterial infections. Picoviruses always kill the cells they infect, after binding to the bacterial cell wall, enzymatically breaking through that wall, penetrating the cell membrane and injecting viral DNA into the cell. They also have other traits that make them attractive candidates for therapeutic use, including a small genome and an inability to transfer bacterial genes between bacteria.
Knowledge of protein structure in Andhra and understanding of how those structures allow the virus to infect a bacterium will make it possible to produce custom-made phages tailored to a specific purpose, using genetic manipulation. "The structural basis for host specificity between phages that infect S. aureus and S. epidermidis is still poorly understood," said Dokland, a professor of microbiology at UAB and director of the UAB Cryo-Electron Microscopy Core. "With the present study, we have gained a better understanding of the structures and functions of the Andhra gene products and the determinants of host specificity, paving the way for a more rational design of custom phages for therapeutic applications. Our findings elucidate critical features for virion assembly, host recognition and penetration." Staphylococcal phages typically have a narrow range of bacteria they can infect, depending on the variable polymers of wall teichoic acid on the surface of different bacterial strains. "This narrow host range is a double-edged sword: On one hand, it allows the phages to target only the specific pathogen causing the disease; on the other hand, it means that the phage may need to be tailored to the patient in each specific case," Dokland said. The general structure of Andhra is a 20-faced, roundish icosahedral capsid head that contains the viral genome. The capsid is attached to a short tail. The tail is largely responsible for binding to S. epidermidis and enzymatically breaking the cell wall. The viral DNA is injected into the bacterium through the tail. Segments of the tail include the portal from the capsid to the tail, and the stem, appendages, knob and tail tip. The 11 different proteins that make up each virus particle are found in multiple copies that assemble together. For instance, the capsid is made of 235 copies each of two proteins, and the other nine virion proteins have copy numbers from two to 72. In total, the virion is made up of 645 protein pieces that include two copies of a 12th protein, whose structure was predicted using the protein structure prediction program AlphaFold.
The atomic models described by Dokland, Hatoum-Aslan, and co-first authors N'Toia C. Hawkins, Ph.D., and James L. Kizziah, Ph.D., UAB Department of Microbiology, show the structures for each protein—as described in molecular language like alpha-helix, beta-helix, beta-strand, beta-barrel or beta-prism. The researchers have described how each protein binds to other copies of that same protein type, such as to make up the hexameric and pentameric faces of the capsid, as well as how each protein interacts with adjacent different protein types. Electron microscopes use a beam of accelerated electrons to illuminate an object, providing much higher resolution than a light microscope. Cryo-electron microscopy adds the element of super-cold temperatures, making it particularly useful for near-atomic structure resolution of larger proteins, membrane proteins or lipid-containing samples like membrane-bound receptors, and complexes of several biomolecules together. In the past eight years, new electron detectors have created a tremendous jump in resolution for cryo-electron microscopy over normal electron microscopy. Key elements of this so-called "resolution revolution" for cryo-electron microscopy are:
- Flash-freezing aqueous samples in liquid ethane cooled to below -256 degrees F. Instead of ice crystals that disrupt samples and scatter the electron beam, the water freezes to a window-like "vitreous ice."
- The sample is kept at super-cold temperatures in the microscope, and a low dose of electrons is used to avoid damage to the proteins.
- Extremely fast direct electron detectors are able to count individual atoms at hundreds of frames per second, allowing sample movement to be corrected on the fly.
- Advanced computing merges thousands of images to generate three-dimensional structures at high resolution. Graphics processing units are used to churn through terabytes of data.
- The microscope stage that holds the sample can also be tilted as images are taken, allowing construction of a three-dimensional tomographic image, similar to a CT scan at the hospital.
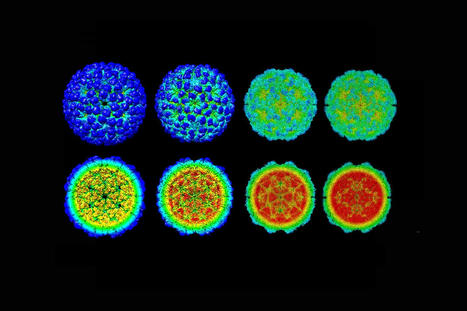
The analysis of Andhra virion structure by the UAB researchers started with 230,714 particle images. Molecular reconstruction of the capsid, tail, distal tail and tail tip started with 186,542, 159,489, 159,489 and 159,489 images, respectively. Resolution ranged from 3.50 to 4.90 angstroms.
Cited research published (Nov. 30, 2022) in Science Advances:



 Your new post is loading...
Your new post is loading...