Your new post is loading...

|
Scooped by
Juan Lama
|
This study presents the first evidence that full vaccination against COVID-19 suppresses emergent mutations of SARS-CoV-2 delta variants. An evolution algorithm, Tajima’s D test with a threshold value as -2.50, can provide a promising tool to forecast new COVID-19 outbreaks. Question It remains unclear how human interventions (vaccinations, lockdowns, etc.) affect viral mutation or generate selection pressure of SARS-CoV-2. It has also been obscure if there are differences in various geographic populations. Findings The vaccination coverage rate is inversely correlated to the mutation frequency of the SARS-CoV-2 delta variants in 16 countries of 20 countries studied. We also discovered delta variants evolved differently under the positive selection pressure in the United Kingdom and India. Meaning Full vaccination against COVID-19 is critical to suppress emergent mutations. Tajima’s D test score, with a threshold value as -2.50, can provide a promising tool to forecast new COVID-19 outbreaks. Available as preprint in medRxiv (August 10, 2021): https://doi.org/10.1101/2021.08.08.21261768

|
Scooped by
Juan Lama
|
A study involving more than 5,000 COVID-19 patients in Houston finds that the virus that causes the disease is accumulating genetic mutations, one of which may have made it more contagious. According to the paper published in the peer-reviewed journal mBIO, that mutation, called D614G, is located in the spike protein that pries open our cells for viral entry. It’s the largest peer-reviewed study of SARS-CoV-2 genome sequences in one metropolitan region of the U.S. to date. The paper shows “the virus is mutating due to a combination of neutral drift — which just means random genetic changes that don’t help or hurt the virus — and pressure from our immune systems,” said Ilya Finkelstein, associate professor of molecular biosciences at The University of Texas at Austin and co-author of the study. The study was carried out by scientists at Houston Methodist Hospital, UT Austin and elsewhere. During the initial wave of the pandemic, 71% of the novel coronaviruses identified in patients in Houston had this mutation. When the second wave of the outbreak hit Houston during the summer, this variant had leaped to 99.9% prevalence. This mirrors a trend observed around the world. A study published in July based on more than 28,000 genome sequences found that variants carrying the D614G mutation became the globally dominant form of SARS-CoV-2 in about a month. SARS-CoV-2 is the coronavirus that causes COVID-19. So why did strains containing this mutation outcompete those that didn’t have it? Perhaps they’re more contagious. A study of more than 25,000 genome sequences in the U.K. found that viruses with the mutation tended to transmit slightly faster than those without it and caused larger clusters of infections. Natural selection would favor strains of the virus that transmit more easily. But not all scientists are convinced. Some have suggested another explanation, called “founder’s effects.” In that scenario, the D614G mutation might have been more common in the first viruses to arrive in Europe and North America, essentially giving them a head start on other strains. The spike protein is also continuing to accumulate additional mutations of unknown significance. The Houston Methodist-UT Austin team also showed in lab experiments that at least one such mutation allows spike to evade a neutralizing antibody that humans naturally produce to fight SARS-CoV-2 infections. This may allow that variant of the virus to more easily slip past our immune systems. Although it is not clear yet whether that translates into it also being more easily transmitted between individuals. The good news is that this mutation is rare and does not appear to make the disease more severe for infected patients. According to Finkelstein, the group did not see viruses that have learned to evade first-generation vaccines and therapeutic antibody formulations. “The virus continues to mutate as it rips through the world,” Finkelstein said. “Real-time surveillance efforts like our study will ensure that global vaccines and therapeutics are always one step ahead.” The scientists noted a total of 285 mutations across thousands of infections, although most don’t appear to have a significant effect on how severe the disease is. Ongoing studies are continuing to surveil the third wave of COVID-19 patients and to characterize how the virus is adapting to neutralizing antibodies that are produced by our immune systems. Each new infection is a roll of the dice, an additional chance to develop more dangerous mutations. Preprint of Study available at medRxiv (Sept. 29, 2020): https://doi.org/10.1101/2020.09.22.20199125

|
Scooped by
Juan Lama
|
Determining whether genetic changes have increased transmission of COVID-19 is surprisingly hard. It’s only a tiny change. At some point early in the pandemic, one of the 30,000 letters in the genome of SARS-CoV-2 changed from an A to a G. Today, that mutation, at position 23,403, has spread around the world. It is found in the vast majority of newly sequenced viruses and has become the center of a burning scientific question: Has the mutation become so common because it helps the virus spread faster? Or is it just coincidence? More than 6 months into the pandemic, the virus’ potential to evolve in a nastier direction—or, if we’re lucky, become more benign—is unclear. In part that’s because it changes more slowly than most other viruses, giving virologists fewer mutations to study. But some virologists also raise an intriguing possibility: that SARS-CoV-2 was already well adapted to humans when it burst onto the world stage at the end of 2019, having quietly honed its ability to infect people beforehand. On average, the coronavirus accumulates about two changes per month in its genome. Sequencing SARS-CoV-2 genomes helps researchers follow how the virus spreads. Most of the changes don’t affect how the virus behaves, but a few may change the disease’s transmissibility or severity. One of the earliest candidates was the wholesale deletion of 382 base pairs in a gene called ORF8, whose function is unknown. First reported by Linfa Wang and others at the Duke-NUS Medical School in Singapore in a March preprint, the deletion has since been reported from Taiwan as well. A deletion in the same gene occurred early in the 2003 severe acute respiratory syndrome (SARS) outbreak, caused by a closely related coronavirus; lab experiments later showed that variant replicates less efficiently than its parent, suggesting the mutation may have slowed the SARS epidemic. Cell culture experiments suggest the mutation does not have the same benign effect in SARS-CoV-2, Wang says, “but there are indications that it may cause milder disease in patients.” The mutation at position 23,403 has drawn the most attention—in part because it changed the virus’ spike, the protein on its surface that attaches to human cells. The mutation changed the amino acid at position 614 of the spike from an aspartic acid (abbreviated D) to a glycine (G), which is why it’s called G614. In a Cell paper this month, Bette Korber and colleagues at Los Alamos National Laboratory showed that G614 has become more common in almost every nation and region they looked at, whereas D614 is virtually gone (see graphic, below). That might be a sign that it’s outcompeted by G614, but it could also be a coincidence. “Any one mutation may rise to very high frequency across the world, just because of random chance,” says Kristian Andersen, a computational biologist at Scripps Research. “This happens all the time.”.....

|
Scooped by
Juan Lama
|
Scientists have created and described more than 3,800 variations of the protein that the new coronavirus uses to latch on to its targets — a feat that reveals which parts of the protein are crucial for binding to human cells. Scientists have created and described more than 3,800 variations of the protein that the new coronavirus uses to latch on to its targets — a feat that reveals which parts of the protein are crucial for binding to human cells. Before SARS-CoV-2 invades a cell, a viral protein called spike fastens tightly to a receptor that sits on the surface of many human cells. Jesse Bloom at the Fred Hutchinson Cancer Research Center in Seattle, Washington, and his colleagues altered a single amino acid at a time in a key segment of spike to produce 3,804 variants of the protein (T. N. Starr et al. Preprint at bioRxiv http://doi.org/dz8r; 2020). Tests showed that many of these variants bind to the receptor at least as well as the protein in the coronavirus causing the current pandemic. The tests allowed the team to pinpoint the amino acids that, if altered, impair the spike protein’s binding ability. This knowledge could help researchers to develop molecules that neutralize the virus’s ability to infect cells. The findings have not yet been peer-reviewed. Preprint available at BioRxiv (June 17, 2020): https://doi.org/10.1101/2020.06.17.157982

|
Scooped by
Juan Lama
|
This is a phylogenetic network of SARS-CoV-2 genomes sampled from across the world. These genomes are closely related and under evolutionary selection in their human hosts, sometimes with parallel evolution events, that is, the same virus mutation emerges in two different human hosts. This makes character-based phylogenetic networks the method of choice for reconstructing their evolutionary paths and their ancestral genome in the human host. The network method has been used in around 10,000 phylogenetic studies of diverse organisms, and is mostly known for reconstructing the prehistoric population movements of humans and for ecological studies, but is less commonly employed in the field of virology.... The described core mutations have been confirmed by a variety of contributing laboratories and sequencing platforms and can be considered reliable. The phylogeographic patterns in the network are potentially affected by distinctive migratory histories, founder events, and sample size. Nevertheless, it would be prudent to consider the possibility that mutational variants might modulate the clinical presentation and spread of the disease. The phylogenetic classification provided here may be used to rule out or confirm such effects when evaluating clinical and epidemiological outcomes of SARS-CoV-2 infection, and when designing treatment and, eventually, vaccines.
|

|
Scooped by
Juan Lama
|
COVID-19 is caused by the coronavirus SARS-CoV-2, which jumped into the human population in late 2019 from a currently uncharacterised animal reservoir. Due to this recent association with humans, SARS-CoV-2 may not yet be fully adapted to its human host. This has led to speculations that SARS-CoV-2 may be evolving towards higher transmissibility. The most plausible mutations under putative natural selection are those which have emerged repeatedly and independently (homoplasies). Here, we formally test whether any homoplasies observed in SARS-CoV-2 to date are significantly associated with increased viral transmission. To do so, we develop a phylogenetic index to quantify the relative number of descendants in sister clades with and without a specific allele. We apply this index to a curated set of recurrent mutations identified within a dataset of 46,723 SARS-CoV-2 genomes isolated from patients worldwide. We do not identify a single recurrent mutation in this set convincingly associated with increased viral transmission. Instead, recurrent mutations currently in circulation appear to be evolutionary neutral and primarily induced by the human immune system via RNA editing, rather than being signatures of adaptation. At this stage we find no evidence for significantly more transmissible lineages of SARS-CoV-2 due to recurrent mutations. SARS-CoV-2 has emerged recently and may still adapt to the human host. Here the authors show that none of the so far identified recurrent mutations in SARS-CoV-2 are significantly associated with increased viral transmission. Published in Nature (Nov. 25, 2020): https://doi.org/10.1038/s41467-020-19818-2

|
Scooped by
Juan Lama
|
Neutralizing antibodies elicited by prior infection or vaccination are likely to be key for future protection of individuals and populations against SARS-CoV-2. Moreover, passively administered antibodies are among the most promising therapeutic and prophylactic anti-SARS-CoV-2 agents. However, the degree to which SARS-CoV-2 will adapt to evade neutralizing antibodies is unclear. Using a recombinant chimeric VSV/SARS-CoV-2 reporter virus, we show that functional SARS-CoV-2 S protein variants with mutations in the receptor binding domain (RBD) and N-terminal domain that confer resistance to monoclonal antibodies or convalescent plasma can be readily selected. Notably, SARS-CoV-2 S variants that resist commonly elicited neutralizing antibodies are now present at low frequencies in circulating SARS-CoV-2 populations. Finally, the emergence of antibody-resistant SARS-CoV-2 variants that might limit the therapeutic usefulness of monoclonal antibodies can be mitigated by the use of antibody combinations that target distinct neutralizing epitopes. Preprint available at bioRxiv (July 22, 2020): https://doi.org/10.1101/2020.07.21.214759

|
Scooped by
Juan Lama
|
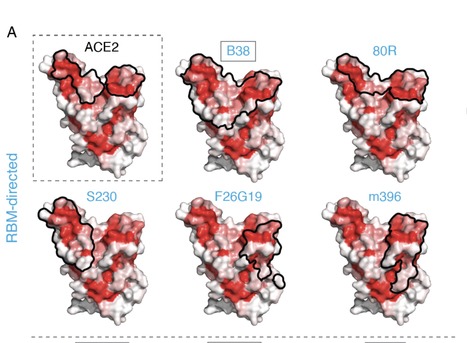
The receptor binding domain (RBD) of the SARS-CoV-2 spike glycoprotein mediates viral attachment to ACE2 receptor, and is a major determinant of host range and a dominant target of neutralizing antibodies. Here we experimentally measure how all amino-acid mutations to the RBD affect expression of folded protein and its affinity for ACE2. Most mutations are deleterious for RBD expression and ACE2 binding, and we identify constrained regions on the RBD’s surface that may be desirable targets for vaccines and antibody-based therapeutics. But a substantial number of mutations are well tolerated or even enhance ACE2 binding, including at ACE2 interface residues that vary across SARS-related coronaviruses. However, we find no evidence that these ACE2-affinity enhancing mutations have been selected in current SARS-CoV-2 pandemic isolates. We present an interactive visualization and open analysis pipeline to facilitate use of our dataset for vaccine design and functional annotation of mutations observed during viral surveillance. Preprint available at bioRxiv (June 17, 2020): https://doi.org/10.1101/2020.06.17.157982

|
Scooped by
Juan Lama
|
The coronavirus is an oily membrane packed with genetic instructions to make millions of copies of itself. The instructions are encoded in 30,000 “letters” of RNA — a, c, g and u — which the infected cell reads and translates into many kinds of virus proteins. In December, a cluster of mysterious pneumonia cases appeared around a seafood market in Wuhan, China. In early January, researchers sequenced the first genome of a new coronavirus, which they isolated from a man who worked at the market. That first genome became the baseline for scientists to track the SARS-CoV-2 virus as it spreads around the world. A cell infected by a coronavirus releases millions of new viruses, all carrying copies of the original genome. As the cell copies that genome, it sometimes makes mistakes, usually just a single wrong letter. These typos are called mutations. As coronaviruses spread from person to person, they randomly accumulate more mutations. The genome below came from another early patient in Wuhan and was identical to the first case, except for one mutation. The 186th letter of RNA was u instead of c. When researchers compared several genomes from the Wuhan cluster of cases they found only a few new mutations, suggesting that the different genomes descended from a recent common ancestor. Viruses accumulate new mutations at a roughly regular rate, so the scientists were able to estimate that the origin of the outbreak was in China sometime around November 2019. Outside of Wuhan, that same mutation in the 186th letter of RNA has been found in only one other sample, which was collected seven weeks later and 600 miles south in Guangzhou, China. The Guangzhou sample might be a direct descendent of the first Wuhan sample. Or they might be viral cousins, sharing a common ancestor. During those seven weeks, the Guangzhou lineage jumped from person to person and went through several generations of new viruses. And along the way, it developed two new mutations: Two more letters of RNA changed to u....
|



 Your new post is loading...
Your new post is loading...