Your new post is loading...

|
Scooped by
Juan Lama
|
By cutting trees in response to international demand for tobacco, farmers induced wildlife to start eating virus-laden bat guano. Zoonotic diseases, or illnesses transmitted from animals to humans, account for about three quarters of new infectious diseases around the world, including some that could lead to pandemics. The risk of a pathogen jumping from an animal to a human increases when people encroach on ecosystems and cause relationships to be disrupted between species—but how that risk actually becomes a reality can be unpredictable and difficult to untangle. A new paper published this week in Communications Biology shines rare light on one such case study: an example showing how international demand for tobacco led to habitat alterations in Uganda that seemingly drove chimpanzees and other species to begin consuming bat guano for mineral nutrients. In that process, the animals might have been exposed to more than two dozen viruses, including a novel cousin of the COVID-causing pathogen SARS-CoV-2...

|
Scooped by
Juan Lama
|
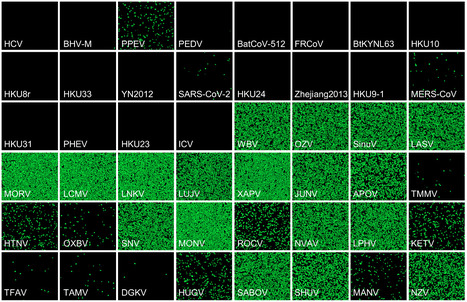
Recent advances in viral metagenomics have led to the discovery of many mammalian viruses, but experimental tests to determine whether they pose a threat to humans are largely lacking. A first step for a virus to cross the species barrier is to penetrate host cells. Here, we use gene synthesis and viral pseudotyping to experimentally test the ability of viral receptor-binding proteins (RBPs) from >100 enveloped RNA viruses to mediate entry into human cells. Analysis of thousands of RBP-cell pairs demonstrated such ability for most viruses, with significant variation among the 14 viral families studied. Comparison of RBP-mediated infectivity with cellular gene expression data showed that viral entry is often not limited by the presence of a receptor and revealed the contribution of additional host factors. Our results prove the weakness of interspecies barriers at the early stages of infection and identify molecular interactions that enable viral zoonosis. Preprint available (Jan. 23, 2024): https://doi.org/10.1101/2024.01.22.576693

|
Scooped by
Juan Lama
|
Horseshoe bats carry viruses closely related to SARS-CoV-2, but they probably can’t spread in people yet. Coronavirus hunters looking for the next pandemic threats have focused on China and southeast Asia, where wild bats carry SARS-CoV-2’s closest known relatives. But a survey of UK bat species suggests that researchers might want to cast a wider net. The trawl turned up new coronaviruses, and some from the same group as SARS-CoV-2. Laboratory studies with safe versions of these viruses suggest that some share key adaptations with SARS-CoV-2 — but are unlikely to spread in humans without further evolution. SARS-CoV-2 belongs to a group of coronaviruses called sarbecoviruses, which circulate in bats. But before the pandemic, efforts to find and characterize these viruses focused on Asia. “Europe and the UK had been totally overlooked,” says Vincent Savolainen, an evolutionary geneticist at Imperial College London who led the study, published on 27 June in Nature Communications. To help plug this gap, Savolainen and his colleagues teamed up with groups involved in bat rehabilitation and conservation to collect a total of 48 faecal samples from bats representing 16 of the 17 species that breed in the United Kingdom. Genetic sequencing turned up nine coronaviruses, including four sarbecoviruses and one related to the coronavirus responsible for Middle East Respiratory Syndrome, or MERS, which periodically spills over into camels and humans. Ties that bind — or not To gauge the threat posed by the UK coronaviruses, the researchers created pseudoviruses: non-replicating forms of HIV that are engineered to carry the spike protein that coronaviruses use to infect cells. One sarbecovirus found in a lesser horseshoe bat (Rhinolophus hipposideros) had a spike protein that was able to infect human cells by attaching to a protein called ACE2, the same receptor used by SARS-CoV-2. But the UK sarbecovirus’s version of spike didn’t attach nearly as strongly as SARS-CoV-2’s, and pseudoviruses could infect only human cells with unnaturally high levels of ACE2. This makes it unlikely that the virus could readily infect people and spread, the researchers say. That’s reassuring, but other sarbecoviruses circulating in British bats could be able to bind to human ACE2 more efficiently, says Michael Letko, a molecular virologist at Washington State University in Pullman, who was not involved in the study. A February preprint surveyed UK lesser horseshoe bats and found signs that around half were infected with sarbecoviruses. Further adaptations that help these viruses to infect human cells more efficiently might not be hard to come by, Letko says. “Once the virus has its foot in the door, it’s easier to adapt further.” Tyler Starr, a molecular evolutionary biologist at the University of Utah in Salt Lake City, says that sarbecoviruses identified so far in Europe and in Africa probably represent the tip of the iceberg when it comes to the group’s true diversity and geographical distribution. He wouldn’t be surprised if the next sarbecovirus to spill over into humans came from an unprecedented location or branch of the family tree. “The next pandemic threat could very well be in our own backyard,” adds Letko. Cited research published in Nat. Communications (June 27, 2023): https://doi.org/10.1038/s41467-023-38717-w

|
Scooped by
Juan Lama
|
Parts of Southeast Asia where human and bat population densities are highest could be infection hotspots, a study finds. Tens of thousands of people in Southeast Asia could be infected with coronaviruses related to SARS-CoV-2 each year, a study published yesterday (August 9) in Nature Communications estimates. The research, which first appeared as a preprint last September, analyzed the geographic ranges of 26 bat species and found their habitat overlapped regions where half a billion people live, representing an area larger than 5 million square kilometers, reports Reuters. Analyzing that data along with estimates of the number of people who exhibited detectable coronavirus antibodies predicted that approximately 66,000 potential infections occur each year. Stephanie Seifert, a virus ecologist at Washington State University in Pullman who was not involved in the research, tells Nature that the work “highlights how often these viruses have the opportunity to spillover.” The study coauthors considered the geographic ranges of bats known to host SARS-related viruses—primarily horseshoe bats (family Rhinolophidae) and Old World leaf-nosed bats (family Hipposideridae). They found hot spots of potential spillover events in southern China, parts of Myanmar, and the Indonesian island of Java—where both bat and human populations are particularly dense, reports Nature. Most of these SARS-related viruses don’t easily spread among humans or cause illness. But study coauthor Peter Daszak tells Nature that with enough infections “raining down on people, you will eventually get a pandemic.” However, Alice Hughes, a conservation biologist at the University of Hong Kong who was not involved in the work, tells Nature that this analysis relies on outdated and low-resolution geographical range data. “What they are trying to do is very valuable and needs to be done, but it has to be done with more finesse,” she says. The study authors argue their research can focus spillover monitoring to high-risk regions in order to identify outbreaks sooner. Renata Muylaert, a disease ecologist at Massey University in New Zealand who was not involved in the research, agrees, telling Nature, “The article has considerable significance for surveillance.” This analysis looked only at bat-to-human spillover events and did not consider infections that first transmit from bats to an intermediate animal and later to humans. Daszak tells Nature that there were limited data on that type of event, but that including them would have “massively increased the estimated risk of spillovers.”

|
Scooped by
Juan Lama
|
The spillover of SARS-CoV-2 into humans has caused one of the most devastating pandemics in recorded history. Human-animal interactions have led to transmission events of SARS-CoV-2 from humans to wild and captive animals. However, many questions remain about how extensive SARS-CoV-2 exposure is in wildlife, the factors that influence wildlife transmission risk, and whether sylvatic cycles can generate novel variants with increased infectivity and virulence. We sampled 18 different wildlife species in the Eastern U.S. and detected widespread exposure to SARS-CoV-2 across wildlife species. Using quantitative reverse transcription polymerase chain reaction and whole genome sequencing, we conclusively detected SARS-CoV-2 in the Virginia opossum and had equivocal detections in six additional species. Species considered human commensals like squirrels, and raccoons had high seroprevalence, ranging between 62%-71%, and sites with high human use had three times higher seroprevalence than low human-use areas. SARS-CoV-2 genomic data from an infected opossum and molecular modeling exposed previously uncharacterized changes to amino acid residues observed in the receptor binding domain (RBD), which predicts improved binding between the spike protein and human angiotensin-converting enzyme (ACE2) compared to the dominant variant circulating at the time of isolation. These mutations were not identified in human samples at the time of collection. Overall, our results highlight widespread exposure to SARS-CoV-2 in wildlife and suggest that areas with high human activity may serve as important points of contact for cross-species transmission. Furthermore, this work highlights the potential role of wildlife in fueling de novo mutations that may eventually appear in humans. Preprint in bioRxiv (Nov. 07, 2022): https://doi.org/10.1101/2022.11.04.515237

|
Scooped by
Juan Lama
|
The henipavirus can cause respiratory symptoms and is related to Nipah and Hendra viruses, but cannot spread easily in people. A new animal virus that can infect people has been identified in eastern China. But scientists say they are not overly concerned because the virus doesn’t seem to spread easily between people, nor is it fatal. The virus, named Langya henipavirus (LayV), can cause respiratory symptoms such as fever, cough and fatigue, and is closely related to two other henipaviruses known to infect people — Hendra virus and Nipah virus. These also cause respiratory infections, and can be fatal. Researchers think LayV is carried by shrews, which might have infected people directly or through an intermediate animal. The virus was described in the New England Journal of Medicine1 on 4 August. Researchers say LayV has infected only 35 people since 2018, and none of the cases seems to be linked. “There is no particular need to worry about this, but ongoing surveillance is critical,” says Edward Holmes, an evolutionary virologist at the University of Sydney in Australia. Regularly testing people and animals for emerging viruses is important to understand the risk of zoonotic diseases — those that can be transmitted from other animals to humans, he says. Large outbreaks of infectious diseases typically take off after a lot of false starts, says Emily Gurley, an infectious-diseases epidemiologist at Johns Hopkins University in Baltimore, Maryland. “If we are actively looking for those sparks, then we are in a much better position to stop or to find something early.” Hospital surveillance The research team that identified LayV did so while monitoring patients at three hospitals in the eastern Chinese provinces of Shandong and Henan between April 2018 and August 2021. Participants were recruited into the study if they had a fever. The team sequenced the LayV genome from a throat swab taken from the first patient identified with the disease, a 53-year-old woman. The virus was named after a town called Langya, in Shandong, where she was from, says co-author Linfa Wang, a virologist at Duke–National University of Singapore Medical School in Singapore. Throughout the study period, the researchers found 35 people who were infected with LayV, mostly farmers, with symptoms ranging from severe pneumonia to a cough. Most patients said in a questionnaire that they had been exposed to an animal within a month of their symptoms appearing. The LayV genome shows that the virus is most closely related to Mojiang henipavirus, which was first isolated in rats in an abandoned mine in the southern Chinese province of Yunnan in 2012. Henipaviruses belong to the Paramyxoviridae family of viruses, which includes measles, mumps and many respiratory viruses that infect people. Several other henipaviruses have been discovered in bats, rats and shrews, from Australia to South Korea and China, but only Hendra, Nipah and now LayV are known to infect people. The researchers did not find strong evidence of LayV spreading between people — there were no clusters of cases in the same family, within a short time span or in close geographical proximity. “Of the 35 cases, not a single one is linked,” says Wang. Gurley says that this is good news, but the study did retrospective contact tracing on only 15 family members of 9 infected individuals, which makes it difficult to determine how exactly the individuals were exposed. Still, she notes that she didn’t see anything in the data to “cause alarm from a pandemic-threat perspective”. Animal origin To determine the potential animal origin of the virus, the researchers tested goats, dogs, pigs and cattle living in the villages of infected patients for antibodies against LayV, and took tissue and urine samples from 25 species of wild small animals to look for the presence of LayV RNA. They found LayV antibodies in a handful of goats and dogs, and identified LayV viral RNA in 27% of the 262 sampled shrews. This suggested that shrews are a reservoir for the virus, passing LayV between themselves “and somehow infecting people here and there by chance”, says Gurley. But it is not clear how people were infected in the first place — whether directly from shrews or an intermediate animal, says Gurley. A lot of research still needs to be done to work out how the virus is spreading in shrews and how people are getting infected, she says. Holmes says there is an urgent need for a global surveillance system to detect virus spillovers and rapidly communicate those results to avoid more pandemics, such as the one sparked by COVID-19. “These sorts of zoonotic spillover events happen all the time,” he says. “The world needs to wake up.” Published in Nature (August 11, 2022): https://doi.org/10.1038/d41586-022-02175-z

|
Scooped by
Juan Lama
|
Abstract A veterinarian in Thailand was diagnosed with COVID-19 after being sneezed on by an infected cat owned by an infected patient. Genetic study supported the hypothesis of SARS-CoV-2 transmission from the owner to the cat, and then from the cat to the veterinarian. Conclusions The identical SARS-CoV-2 genome sequences obtained from patient A and the sequences derived from the cat and its 2 owners, together with the temporal overlapping of the animal and human infections, indicated that their infections were epidemiologically related. Because patient A had no prior meetings with patients B or C, she probably acquired SARS-CoV-2 from the cat when it sneezed in her face. The genome sequences were distinct from that of patient G and other sequences circulating in the same province, and by using the pairwise distance formula, we were able to rule out external transmission (5). The Alpha variant was widely spread until the end of July 2021 in Songkhla Province; on the other hand, in Bangkok, the Delta variant has been widespread since the beginning of July 2021 (6). The transmission chain of SARS-CoV-2 infections in this cluster probably began in Bangkok. Cats are known to be susceptible to SARS-CoV-2 infection (8–10), especially during close interactions with humans with symptomatic SARS-CoV-2 infections (7). Because infected cats have relatively short incubation and contagious periods (8–10), this cat probably had acquired its SARS-CoV-2 infection no longer than a week before possibly transmitting the disease to patient A. Although direct or indirect (fomites) contacts are also potential routes of transmission to patient A, these possibilities are less likely because she wore gloves and washed her hands before and after examining the cat. Transmission from the cat sneeze is hypothesized because of this brief but very close encounter. The relatively low RT-PCR cycle thresholds (11) in the nasal swab obtained from the cat suggest that the viral load was high and infectious (12,13). Because patient A wore an N95 mask without a face shield or goggles, her exposed ocular surface was vulnerable to infection by droplets expelled from the cat. Her infection signifies the possibility of ocular transmission and the importance of wearing protective goggles or face shields in addition to a mask during close-range interactions with high-risk humans or animals. In summary, we have provided evidence that cats can transmit the SARS-CoV-2 infection to humans. However, the incidence of this transmission method is relatively uncommon because of the short (median 5 days) duration of cats shedding viable viruses (8–10). Nevertheless, to prevent transmission of SARS-CoV-2 from humans to cat, persons with suspected or confirmed COVID-19 should refrain from contact with their cat. Eye protection as part of the standard personal protection is advisable for caregivers during close interactions with cats suspected to be infected. Top Mr. Sila is a graduate student at Prince of Songkla University’s Department of Health Science and Medical Research in the Faculty of Medicine. His primary research interests include genomics, evolutionary microbiology, bioinformatics, sequence and genome analysis, and viral culture. Published in Emerging Infectious Diseases: 10.3201/eid2807.212605

|
Scooped by
Juan Lama
|
A core question in understanding the drivers of zoonotic emergence is whether particular animal groups are common sources of zoonotic viruses. If so, can this information be used to establish a watch-and-act list of those species most likely to carry potentially pandemic viruses? It has long been known that most viral infections in humans have their ancestry in mammals (4). Birds are the only other probable source of zoonotic diseases, with the various forms of avian influenza virus that occasionally appear in humans (such as H5 subtype viruses) presenting an ongoing pandemic threat. Although viruses are often abundant in other animal groups (such as bony fish), their phylogenetic distance to humans greatly reduces the likelihood of successful cross-species transmission. Within the mammals, a variety of groups have served as reservoirs for zoonotic viruses (3), particularly those with which humans share proximity, either as food sources (such as pigs) or because they have adapted to human lifestyles (like some species of rodent), as well as those that are so closely related to humans (such as nonhuman primates) that viruses face little adaptive challenge when establishing human-to-human transmission. Most of all, since the emergence of SARS in late 2002, there has been intense interest in bats as virus reservoirs, although this may in part reflect biases in both ascertainment and confirmation (5). Although bats seemingly tolerate a high diversity and abundance of viruses, the underlying immunological, physiological, and ecological reasons for this are not fully understood (6). More pragmatically, the majority of bat viruses have not appeared in humans, and those that have emerged often do so through other host species (i.e., “intermediate hosts”) prior to successful emergence (7). Bats are important players in disease emergence, but they are only one component of the more complex global viral ecosystem. A related question is whether the viruses that are most likely to emerge in humans can be identified. Although metagenomic sequencing is revealing an increasingly large virosphere, with mammals carrying many thousands of different viruses, most of which remain undocumented (5), the greatest pandemic risk is posed by respiratory viruses because their fluid mode of (sometimes asymptomatic) transmission makes their control especially challenging. Three groups of RNA viruses that regularly jump species boundaries best fit this risk profile: paramyxoviruses, influenza viruses, and, particularly, coronaviruses. Hendra and Nipah viruses, both with ultimate bat ancestry, are exemplars of paramyxoviruses that have emerged in humans. Although neither have resulted in large-scale outbreaks, it is possible that more transmissible paramyxoviruses (such as the case of measles virus) lurk in the mammalian virosphere. The documented host range of influenza viruses is growing, including recent reports of avian H9N2 influenza virus in diseased Asian badgers (2), but most human influenza virus pandemics have their roots in those viruses that circulate in waterbirds and poultry, often with the secondary involvement of pigs. Fortunately, birds and humans are sufficiently different in most virus–cell interactions that avian viruses are usually unable to successfully transmit among humans......

|
Scooped by
Juan Lama
|
Scientists find relatives of rubella in bats, wild mice, and zoo animals. The virus that causes rubella, or German measles, finally has company. Scientists had never identified close relatives of the virus, leaving it as the only member of its genus, Rubivirus. But with a report in this week’s issue of Nature, rubella has gained a family. One of its two newfound relatives infects bats in Uganda; the other killed animals from three different species in a German zoo and was found in wild mice living nearby as well. The findings strongly suggest that at some point in the past, a similar virus jumped from animals to humans, giving rise to today’s rubella virus, the researchers say. Although neither of the new viruses is known to infect humans, the fact that a related virus jumped species raises concerns that the two viruses or other, as-yet-unknown relatives could cause human outbreaks. “We would be remiss not to be concerned, given what’s going on in the world today,” says epidemiologist Tony Goldberg of the University of Wisconsin, Madison, a senior author of the study. Highly infectious, the rubella virus usually causes rashes and fever, but in pregnant women it can lead to miscarriages, stillbirth, and babies born with congenital rubella syndrome, which includes deafness and eye, heart, and brain problems. An estimated 100,000 newborns are affected by the syndrome annually, mostly in Africa, the western Pacific, and the eastern Mediterranean; in many other countries the measles, mumps, and rubella (MMR) vaccine has made it a rarity. Goldberg and his former graduate student Andrew Bennett discovered one of the new viruses in apparently healthy cyclops leaf-nosed bats, netted at night in Kibale National Park in Uganda. They named it ruhugu virus, after the Ruteete region of Uganda and the local word for bat. The architecture of ruhugu’s genome is identical to that of the rubella virus, and 56% of the amino acids in its eight proteins matched those in rubella. The protein that interacts with the host’s immune cells was almost identical in both viruses. As they were getting ready to publish, the researchers learned that a team led by Martin Beer at the Friedrich-Loeffler Institute had detected another rubella relative in brain tissue from a donkey, a kangaroo, and a capybara—a giant rodent native to South America—that all died from encephalitis, an inflammation of the brain, at an unnamed zoo. They found the same virus in wild yellow-necked field mice caught in the zoo or within a 10-kilometer radius. The mice appeared to be fine, suggesting they were a natural reservoir from which the virus spilled over to the zoo animals. Comparing their data, the teams realized their viruses were related, although ruhugu was closer to rubella than the second relative, rustrela virus, named after a lagoon in the Baltic Sea. The teams decided to publish jointly. Two other viruses that primarily affect children, measles and mumps, also came from animals, Goldberg notes. “Now we know that every disease in the letters of the MMR vaccine has a zoonotic origin,” he says. Given the genetic distance between rubella and the ruhugu and rustrela viruses, the researchers don’t think either of them made the jump to humans—but they suspect they’ll find other Rubiviruses if they look closely. The paper is “really important because there’s very little understanding of where rubella came from,” says molecular anthropologist Anne Stone of Arizona State University, Tempe. “It was all by itself without any close relative.” The finding underscores the importance of the One Health approach, which recognizes that the health of people is closely connected to that of animals and the environment, she says. Both viruses bear close watching, researchers say. It’s “really interesting” that rustrela was able to infect both placental and marsupial mammals, and “was actively jumping between species,” says evolutionary virologist Edward Holmes of the University of Sydney. That flexibility could spell trouble, says vaccinologist Gregory Poland of the Mayo Clinic. “Who knows, if it could move from mice to other mammals, could it move to humans?” he asks. “In the end, the bugs win.” Study published in Nature (October 7, 2020): https://doi.org/10.1038/s41586-020-2812-9
|

|
Scooped by
Juan Lama
|
Newswise — Retroviruses are viruses that multiply by incorporating their genes into the genome of a host cell. If the infected cell is a germ cell, the retrovirus can then be passed on to the next generation as an “endogenous” retrovirus (ERV) and spread as part of the host genome in that host species. In vertebrates, ERVs are ubiquitous and sometimes make up 10 per cent of the host genome. However, most retrovirus integrations are very old, already degraded and therefore inactive – their initial impact on host health has been minimised by millions of years of evolution. A research team led by the Leibniz Institute for Zoo and Wildlife Research (Leibniz-IZW) has now discovered a recent case of retrovirus colonisation in a rodent from New Guinea, the white-bellied mosaic-tailed rat. In a paper in the scientific journal "Proceedings of the National Academy of Sciences", they describe this new model of virus integration. The observations on this process will help to improve our understanding how retroviruses rewrite host genomes.
Retroviruses, such as the pathogen responsible for AIDS (HIV-1), integrate into the genome of the host cells they infect during their life cycle. When this happens in the germline (egg cells or cells that produce sperm) of the host, the retrovirus can actually become a gene of the host itself. This process is apparently common, as up to 10 percent of the genomes of most vertebrates consist of the remnants of such ancient infections. One of the best studied models of this process is the koala retrovirus (KoRV), which is currently colonising the koala genome. „What happens to the virus and the host during this process of genome colonisation we do not know, as most such events occurred millions of years ago and we only see the leftover ‘fossils’ of the retrovirus”, says Prof Alex Greenwood, head of the Leibniz-IZW Department of Wildlife Diseases. “Nor do we know what the host suffered health-wise during the infection process. The koala retrovirus (KoRV) is one of the few models of this process that occurs in real time and where we can observe the effects of genome colonisation on the host animal.”
There is now some evidence that KoRV-related viruses are circulating in rodents and bats in Papua New Guinea and Indonesia. A group led by Greenwood and Dr Saba Mottaghinia, former PhD student in Greenwood’s department, analysed 278 samples from seven bat and one rodent family endemic to the Australo-Papua region (Australia and New Guinea). They discovered a retrovirus that is currently colonising the genome of an endemic rodent from New Guinea, the white-bellied mosaic-tailed rat (Melomys leucogaster). This is only the second example from the Australo-Papuan region, after KoRV, of a retrovirus that has colonised a genome while retaining a functional viral life cycle. The gibbon ape leukaemia viruses (GALV), a group of viruses discovered in gibbons and woolly monkeys at a research facility in Thailand in the 1960s, are very closely related to KoRV. This is a curious and surprising relationship, as there is a geographical barrier, known as the Wallace lineage, which separates the fauna of Southeast Asia from Indonesia, Papua New Guinea and Australia's fauna. However, there is evidence that the gibbons and woolly monkeys at the research facility in Thailand have been infected with viruses from Papua New Guinea. „The discovery of GALV-like viruses in rodents and bats in Indonesian and Australian rodents and bats from New Guinea suggests that these viruses, and possibly also KoRV, originated in New Guinea”, says Greenwood, who initiated the research project funded by the German Research Foundation.
The Leibniz-IZW team, together with scientists from the Charité, the Robert Koch Institute, the Max Delbrück Center, the University of Nicosia, California State University Fullerton, the South Australian Museum and Museum Victoria, examined 278 bat and rodent samples from Australia and New Guinea for KoRV and GALV-like viruses. They detected a GALV, the Woolly Monkey Virus (WMV) in a population of the white-bellied mosaic-tailed rat, an endemic rodent from New Guinea. In five of the rats from two New Guinea collection sites, WMV was integrated into the genome at the same location, indicating that it has spread as a gene and not by infection, i.e. it has become part of the genome. However, in other white-bellied mosaic-tailed rat populations the virus was absent – similar to KoRV in koalas, where all koalas in northern Australia have KoRV in their genome, whereas there are koalas in southern Australia that do not have intact KoRV. The virus, now called the “complete Melomys woolly monkey virus” (cMWMV), was able to infect cell lines, produce new viral progeny, was visible by electron microscopy as viral particles that detached from the cell membrane, and was even sensitive to the antiretroviral drug AZT.
„The virus has all the characteristics of an exogenous infectious retrovirus, but is endogenous. It is probably a very recent colonisation event, much younger than KoRV", says Dr Saba Mottaghinia, the lead author of the paper in the „Proceedings of the National Academy of Sciences“. The results suggest that cMWMV is a new model for retroviral colonisation of the host genome that occurs in real time, as in KoRV, and they also suggest that GALVs like WMV originated in the diverse fauna of New Guinea. The discoveries in New Guinea have certainly not been exhausted. “There are hundreds of mammalian species from this region that have not yet been studied, suggesting that many more viruses and models exist in this region”, says Greenwood.
The authors dedicate the study to Ken P. Aplin of the South Australian Museum, who sadly passed away during the course of the project
Published in PNAS (Feb. 1, 2024): https://doi.org/10.1073/pnas.2220392121

|
Scooped by
Juan Lama
|
Study finds deer are virus reservoirs, promoting ongoing mutation. New research has found that white-tailed deer across Ohio have been infected with the virus that causes COVID-19. Alarmingly, the results also show that viral variants evolve about three times faster in deer than in humans. Scientists collected 1,522 nasal swabs from free-ranging deer in 83 of the state’s 88 counties between November 2021 and March 2022. More than 10% of the samples were positive for the SARS-CoV-2 virus, and at least one positive case was found in 59% of the counties in which testing took place. Genomic analysis showed that at least 30 infections in deer had been introduced by humans – a figure that surprised the research team. “We generally talk about interspecies transmission as a rare event, but this wasn’t a huge sampling, and we’re able to document 30 spillovers. It seems to be moving between people and animals quite easily,” said Andrew Bowman, associate professor of veterinary preventive medicine at The Ohio State University and co-senior author of the study. “And the evidence is growing that humans can get it from deer – which isn’t radically surprising. It’s probably not a one-way pipeline.” The combined findings suggest that the white-tailed deer species is a reservoir for SARS-CoV-2 that enables continuing mutation, and that the virus’s circulation in deer could lead to its spread to other wildlife and livestock. The study is published today (August 28, 2023) in the journal Nature Communications. Previous Observations and Expansions Bowman and colleagues previously reported detection of SARS-CoV-2 infections in white-tailed deer in nine Ohio locations in December 2021, and are continuing to monitor deer for infection by more recent variants. “We expanded across Ohio to see if this was a localized problem – and we find it in lots of places, so it’s not just a localized event,” Bowman said. “Some of the thought back then was that maybe it’s just in urban deer because they’re in closer contact with people. But in rural parts of the state, we’re finding plenty of positive deer.” Beyond the detection of active infections, researchers also found through blood samples containing antibodies – indicating previous exposure to the virus – that an estimated 23.5% of deer in Ohio had been infected at one time or another. Variant Analysis The 80 whole-genome sequences obtained from the collected samples represented groups of various viral variants: the highly contagious delta variant, the predominant human strain in the United States in the early fall of 2021 that accounted for almost 90% of the sequences, and alpha, the first named variant of concern that had circulated in humans in the spring of 2021. The analysis revealed that the genetic composition of delta variants in deer matched dominant lineages found in humans at the time, pointing to the spillover events, and that deer-to-deer transmission followed in clusters, some spanning multiple counties. “There’s probably a timing component to what we found – we were near the end of a delta peak in humans, and then we see a lot of delta in deer,” Bowman said. “But we were well past the last alpha detection in humans. So the idea that deer are holding onto lineages that have since gone extinct in humans is something we were worried about.” The study did suggest that COVID-19 vaccination is likely to help protect people against severe disease in the event of a spillover back to humans. An analysis of the effects of deer variants on Siberian hamsters, an animal model for SARS-CoV-2 studies, showed that vaccinated hamsters did not get as sick from infection as unvaccinated animals. Rapid Evolution in Deer Disturbingly, the variants circulating in deer are expected to continue to change. An investigation of the mutations found in the samples provided evidence of more rapid evolution of both alpha and delta variants in deer compared to humans. “Not only are deer getting infected with and maintaining SARS-CoV-2, but the rate of change is accelerated in deer – potentially away from what has infected humans,” Bowman said. How the virus is transmitted from humans to white-tailed deer remains a mystery. And so far, even with about 30 million free-ranging deer in the U.S., no substantial outbreaks of deer-origin strains have occurred in humans. Potential Implications Circulation among animals, however, remains highly likely. Bowman noted that about 70% of free-ranging deer in Ohio have not been infected or exposed to the virus, “so that’s a large body of naive animals that the virus could spread through rather uninhibited.” “Having that animal host in play creates things we need to watch out for,” he said. “If this trajectory continues for years and we have a virus that becomes deer-adapted, then does that become the pathway into other animal hosts, wildlife or domestic? We just don’t know.” Martha Nelson of the National Library of Medicine was co-corresponding author of the study. Ohio State co-authors Dillon McBride, Steven Overend, Devra Huey, Amanda Williams, Seth Faith and Jacqueline Nolting worked on the study with co-authors from St. Jude Children’s Research Hospital; the University of California, Los Angeles; the National Research Centre in Giza, Egypt; PathAI Diagnostics; the Ohio Department of Natural Resources; the U.S. Department of Agriculture; Columbus and Franklin County Metroparks; and the Rega Institute for Medical Research in Belgium. Cited Study published in Nat. Communications (Aug. 28, 2023) https://doi.org/10.1038/s41467-023-40706-y

|
Scooped by
Juan Lama
|
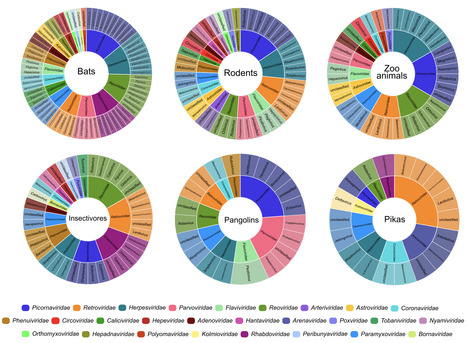
Wildlife is reservoir of emerging viruses. Here we identified 27 families of mammalian viruses from 1981 wild animals and 194 zoo animals collected from south China between 2015 and 2022, isolated and characterized the pathogenicity of eight viruses. Bats harbor high diversity of coronaviruses, picornaviruses and astroviruses, and a potentially novel genus of Bornaviridae. In addition to the reported SARSr-CoV-2 and HKU4-CoV-like viruses, picornavirus and respiroviruses also likely circulate between bats and pangolins. Pikas harbor a new clade of Embecovirus and a new genus of arenaviruses. Further, the potential cross-species transmission of RNA viruses (paramyxovirus and astrovirus) and DNA viruses (pseudorabies virus, porcine circovirus 2, porcine circovirus 3 and parvovirus) between wildlife and domestic animals was identified, complicating wildlife protection and the prevention and control of these diseases in domestic animals. This study provides a nuanced view of the frequency of host-jumping events, as well as assessments of zoonotic risk. Monitoring the diversity of viruses infecting animals is important for assessing zoonotic risk. Here, the authors use metatranscriptomics to characterise the viromes of small mammals, pangolins, and zoo animals in China to identify potentially zoonotic viruses. Published in Nature Comm. (April 29, 2023): https://doi.org/10.1038/s41467-023-38202-4

|
Scooped by
Juan Lama
|
Landmark study reveals ‘spillover’ mechanism for the rare but deadly Hendra virus. “Hey guys, could you open your wings and show me?” says Peggy Eby, looking up at a roost of flying foxes in Sydney’s Botanical Gardens. “I talk to them a lot.” Eby, a wildlife ecologist at the University of New South Wales in Sydney, Australia, is looking for lactating females and their newborn pups, but the overcast weather is keeping them snuggled under their mothers’ wings. Eby has been studying flying foxes, a type of bat, for some 25 years. Using her binoculars, she tallies the number of lactating females that are close to weaning their young — a proxy for whether the bats are experiencing nutritional stress and so probably more likely to shed viruses that can make people ill. Australian flying foxes are of interest because they host a virus called Hendra, which causes a very rare but deadly respiratory infection that kills one in every two infected people. Hendra virus, like Nipah, SARS-CoV and SARS-CoV-2 (the virus that caused the COVID-19 pandemic) is a bat virus that has spilled over into people. These viruses often reach humans through an intermediate animal, sometimes with deadly consequences. Scientists know that spillovers are associated with habitat loss, but have struggled to pinpoint the specific conditions that spark events until now. After a detailed investigation, Eby and her colleagues can now predict — up to two years ahead — when clusters of Hendra virus spillovers will probably appear. “They have identified the environmental drivers of spillover,” says Emily Gurley, an infectious-diseases epidemiologist at Johns Hopkins University in Baltimore, Maryland. And they have determined how those events could be prevented. The results are published in Nature on 16 November1. Food stress Specifically, the researchers found that clusters of Hendra virus spillovers occur following years in which the bats experience food stress. And these food shortages typically follow years with a strong El Niño, a climatic phenomenon in the tropical Pacific Ocean that is often associated with drought along eastern Australia. But if the trees the bats rely on for food during the winter have a large flowering event the year after there’s been a food shortage, there are no spillovers. Unfortunately, the problem is that “there’s hardly any winter habitat left”, says Raina Plowright, a disease ecologist and study co-author at Cornell University in Ithaca, New York. The study is “absolutely fantastic”, says Sarah Cleaveland, a veterinarian and infectious-disease ecologist at the University of Glasgow, UK. “What’s so exciting about it is that it has led directly to solutions.” Cleaveland says the study’s approach of looking at the impact of climate, environment, nutritional stress and bat ecology together could bring new insights to the study of other pathogens, including Nipah and Ebola, and their viral families. The study provides “a much clearer understanding of drivers of this issue, with broad relevance to pandemics elsewhere”, says Alice Hughes, a conservation biologist at the University of Hong Kong. “The paper underscores the enhanced risk we are likely to see” with climate change and increasing habitat loss, she says. Urban shift Hendra virus was identified in 1994, following an outbreak in horses and people at a thoroughbred training facility in Brisbane, Australia. Studies later established that the virus spreads from its bat reservoir — most likely the black flying fox (Pteropus alecto) — to horses through faeces, urine and spats of chewed-up pulp the flying foxes spit out on the grass. Infected horses then spread the virus to people. Infections typically occur in clusters during the Australian winter, and several years can go by before another cluster emerges in horses, but cases have been picking up since the early 2000s. To study the mechanism of spillovers, Plowright, Eby and their colleagues collected data on the location and timing of such events, the location of bat roosts and their health, climate, nectar shortages and habitat loss over some 300,000-square kilometres in southeast Australia from 1996 to 2020. Then they used modelling to determine which factors were associated with spillovers. “I’m just in awe of the invaluable data sets that they have on the ecology,” says Gurley. Over the course of the study, the team noticed significant changes in the bats’ behaviour. The flying foxes went from having predominantly nomadic lifestyles — moving in large groups from one native forest to the other in search of nectar — to settling in small groups in urban and agricultural areas, bringing the bats closer to where horses and people live. The number of occupied bat roosts in general has trebled since the early 2000s to around 320 in 2020. A separate study from the team2 found that the newly established roosts shed Hendra virus every winter, but in years following a food shortage bats shed more virus. There were “really dramatic winter spikes in infection”, says co-author Daniel Becker, an ecologist who focuses on infectious diseases at the University of Oklahoma in Norman. The study also linked increased viral shedding in bats to increased spillovers to horses. In search of nectar Modelling in Plowright and Eby’s most recent Nature paper shows that flying-fox populations split into small groups that migrated to agricultural areas close to horses when food was scarce, and that food shortages followed strong El Niño events, probably because native eucalyptus tree budding is sensitive to climate changes. To conserve energy, the bats fly only small distances in these years, scavenging for food in agricultural areas near horses. Spillovers to horses were most likely to occur in winters following a food shortage, says Plowright. Their model was able to accurately predict in which years these would occur. Then something unexpected happened. An El Niño occurred in 2018 followed by a drought in 2019, suggesting that 2020 should also have been a spillover year. But there was only one event in May and none has been detected since. “We threw all the cards back up into the air and looked carefully at all the other elements of our hypothesis,” says Eby. Eventually they discovered that when native forests have major flowering events in winters following a food shortage, this helps to avert spillovers. In 2020, a red-gum forest near the town of Gympie flowered, drawing in some 240,000 bats. And similar winter flowering events occurred in other regions in 2021 and 2022. The researchers suggest that these mass migrations take the bats away from horses. They propose that by restoring the habitats of those handful of species that flower in winter, fewer spillovers in horses, and potentially in people, would occur. And by restoring the habitats of other animals that host dangerous pathogens, “maybe we can prevent the next pandemic”, says Plowright. Published in Nature: https://doi.org/10.1038/d41586-022-03682-9

|
Scooped by
Juan Lama
|
The virus that causes COVID-19 is a prolific sack of genes that targets not just humans but nonhuman animals as well. And just as humans and animals can infect one another, animals can also infect other animals, says Amélie Desvars-Larrive, an epidemiologist at the University of Veterinary Medicine in Vienna. Scientists have learned a lot about how COVID spreads in humans but less about how it spreads between animals. To make it easier to study the connections among humans, animals and the virus, Desvars-Larrive and a team of researchers gathered scattered reports of COVID-infected mammals from all over the world to create a public database. Understanding how the virus spreads between nonhuman mammals, and then between those mammals and humans, can help us better manage the current pandemic—and prepare for the next one. “We can’t continue to focus on humans, to have an anthropocentric point of view on this pandemic,” Desvars-Larrive says. COVID has proved highly contagious among many mammal species: Infections have run rampant among captive mink, and fur farmers have had to kill their entire stocks to stop it. Deer appear particularly susceptible to the virus. Many cat species, both big and small, seem to get it as well. Barbara Han, a disease ecologist at the Cary Institute of Ecosystem Studies in Millbrook, N.Y., who was not involved in the database project, says having this kind of information in one place—rather than split among multiple sources managed by different organizations and government agencies—will likely make it faster and easier for her team to predict how the virus behaves. The database is growing as more animals are tested and reports shared, and scientists hope it will help them to track animal-to-animal COVID infections as well as transmission between animals and humans. Han says that beyond just tallying individual infected species, the database will make it easier for scientists to study how the COVID-causing SARS-CoV-2 virus affects mammal communities and entire ecosystems. “People are fascinated that this pathogen is now in [so many] animals and what it might mean for us,” Han says. “If we don’t have good information about which animals have it now, we can’t get those answers.” Database at https://vis.csh.ac.at/sars-ani/

|
Scooped by
Juan Lama
|
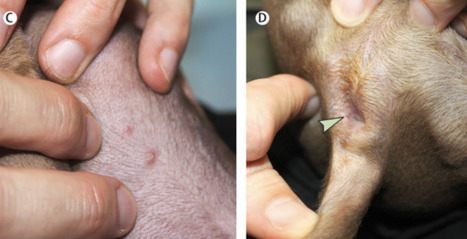
Human monkeypox virus is spreading in Europe and the USA among individuals who have not travelled to endemic areas. On July 23, 2022, monkeypox was declared a Public Health Emergency of International Concern by WHO Director-General Tedros Adhanom Ghebreyesus. Human-to-human transmission of monkeypox virus usually occurs through close contact with the lesions, body fluids, and respiratory droplets of infected people or animals. The possibility of sexual transmission is being investigated, as the current outbreak appears to be concentrated in men who have sex with men and has been associated with unexpected anal and genital lesions. Whether domesticated cats and dogs could be a vector for monkeypox virus is unknown. Here we describe the first case of a dog with confirmed monkeypox virus infection that might have been acquired through human transmission. Two men who have sex with men attended Pitié-Salpêtrière Hospital, Paris, France, on June 10, 2022 (appendix). One man (referred to as patient 1 going forward) is Latino, aged 44 years, and lives with HIV with undetectable viral loads on antiretrovirals; the second man (patient 2) is White, aged 27 years, and HIV-negative. The men are non-exclusive partners living in the same household. They each signed a consent form for the use of their clinical and biological data, and for the publication of anonymised photographs. The men had presented with anal ulceration 6 days after sex with other partners. In patient 1, anal ulceration was followed by a vesiculopustular rash on the face, ears, and legs; in patient 2, on the legs and back. In both cases, rash was associated with asthenia, headaches, and fever 4 days later. 12 days after symptom onset, their male Italian greyhound, aged 4 years and with no previous medical disorders, presented with mucocutaneous lesions, including abdomen pustules and a thin anal ulceration (figure C, D; appendix). The dog tested positive for monkeypox virus by use of a PCR protocol adapted from Li and colleagues that involved scraping skin lesions and swabbing the anus and oral cavity. Monkeypox virus DNA sequences from the dog and patient 1 were compared by next-generation sequencing (MinION; Oxford Nanopore Technologies, Oxford, UK). Both samples contained virus of the hMPXV-1 clade, lineage B.1, which has been spreading in non-endemic countries since April, 2022, and, as of Aug 4, 2022, has infected more than 1700 people in France, mostly concentrated in Paris, where the dog first developed symptoms. Moreover, the virus that infected patient 1 and the virus that infected the dog showed 100% sequence homology on the 19·5 kilobase pairs sequenced. The men reported co-sleeping with their dog. They had been careful to prevent their dog from contact with other pets or humans from the onset of their own symptoms (ie, 13 days before the dog started to present cutaneous manifestations). In endemic countries, only wild animals (rodents and primates) have been found to carry monkeypox virus. However, transmission of monkeypox virus in prairie dogs has been described in the USA and in captive primates in Europe that were in contact with imported infected animals. Infection among domesticated animals, such as dogs and cats, has never been reported. Published in The Lancet (August 10, 2022):

|
Scooped by
Juan Lama
|
A new technology aims to stop wildlife from spreading Ebola, rabies, and other viruses. It could prevent the next pandemic by stopping pathogens from jumping from animals to people. Imagine a cure that’s as contagious as the disease it fights—a vaccine that could replicate in a host’s body and spread to others nearby, quickly and easily protecting a whole population from microbial attacks. That’s the goal of several teams around the world who are reviving controversial research to develop self-spreading vaccines. Their hope is to reduce infectious disease transmission among wild animals, thereby lowering the risk that harmful viruses and bacteria can jump from wildlife to humans as many experts believe happened with SARS-CoV-2, the virus that caused the COVID-19 pandemic. The U.S. Centers for Disease Control and Prevention estimates that 60 percent of all known infectious diseases and 75 percent of new or emerging infectious diseases are zoonotic. Scientists cannot predict why, when, or how new zoonotic diseases will emerge. But when they do, these diseases are often deadly and costly to control. What’s more, many researchers predict that climate change, biodiversity loss, and population growth will accelerate their spread. Vaccines are a key tool for preventing diseases from spreading, but wild animals are difficult to vaccinate because each one must be located, captured, vaccinated, and released. Self-spreading vaccines offer a solution. Advances in genomic technology and virology, and a better understanding of disease transmission, have accelerated work that began in the 1980s to make genetically engineered viruses that spread from one animal to another, imparting immunity to disease rather than infection. Researchers are currently developing self-spreading vaccines for Ebola, bovine tuberculosis, and Lassa fever, a viral disease spread by rats that causes upward of 300,000 infections annually in parts of West Africa. The approach could be expanded to target other zoonotic diseases, including rabies, West Nile virus, Lyme disease, and the plague. Advocates for self-spreading vaccines say they could revolutionize public health by disrupting infectious disease spread among animals before a zoonotic spillover could occur—potentially preventing the next pandemic. But others argue that the viruses used in these vaccines could themselves mutate, jump species, or set off a chain reaction with devastating effects across entire ecosystems. “Once you set something engineered and self-transmissible out into nature, you don't know what happens to it and where it will go,” says Jonas Sandbrink, a biosecurity researcher at the University of Oxford’s Future of Humanity Institute. “Even if you just start by setting it out into animal populations, part of the genetic elements might find their way back into humans.” The first, and only, self-spreading vaccine field trial In 1999, veterinarian José Manuel Sánchez-Vizcaíno led a team of researchers to Isla del Aire, an island off the eastern coast of Spain, to test a self-spreading vaccine against two viral diseases: rabbit hemorrhagic disease and myxomatosis. Although neither disease infects humans, at the time they both had been decimating domestic and wild rabbit populations across China and Europe for several decades. Traditional vaccines for both diseases were used in domestic rabbits, but trapping and vaccinating wild rabbits, which are notoriously fast-breeding, was an insurmountable task, Sánchez-Vizcaíno explains. He saw huge potential in self-spreading vaccines. In the laboratory, Sánchez-Vizcaíno, then the director of the Center for Research in Animal Health in Spain, and his team sliced out a gene from the rabbit hemorrhagic disease virus and inserted it into the genome of a mild strain of the myxoma virus, which causes myxomatosis. The final product was a hybrid virus vaccine that protected against both rabbit hemorrhagic disease and myxomatosis. Sánchez-Vizcaíno hypothesized that because the vaccine was similar enough to the original disease-causing myxoma virus, it would still spread among wild rabbits. On the island the research team captured 147 rabbits, placed microchips in their necks, administered the vaccine to about half of them, and released them all back into the wild. For the next 32 days, the vaccinated and unvaccinated rabbits lived as they normally did. When researchers recaptured micro-chipped rabbits that had not been vaccinated originally, they found that 56 percent of them had antibodies to both viruses, indicating that the vaccine had successfully spread from vaccinated to unvaccinated animals. The experiment was the first proof-of-concept field test for self-spreading vaccines—and it remains the only one ever attempted. In 2000, the research team submitted their laboratory and field data to the European Medicines Agency, or EMA, for evaluation and approval for real-world use. The EMA noted technical issues with the vaccine’s safety evaluation and requested that the team decode the myxoma genome, which had not been done before. Although the team was given two years to comply, the funding body did not provide support for further work, recalls Juan Bárcena, then a Ph.D. student working under Sánchez-Vizcaíno. Bárcena no longer advocates for self-spreading vaccine technology, but he says that data from the laboratory and field trials showed the vaccine was safe and its spread remained confined to the rabbit populations. Still, Bárcena doubts that the EMA would have ever approved their vaccine given the hesitancy and controversy around genetically modified organisms. Scott Nuismer, a professor at the University of Idaho who conducts mathematical modelling studies of self-spreading vaccines today, noted that Sánchez-Vizcaíno’s vaccine may have posed more risks than current technologies because the team used a myxoma virus, which is itself deadly, as its vehicle for the vaccine. After the Isla del Aire field trials, research into self-spreading vaccines went largely dormant. Pharmaceutical companies weren’t interested in investing in research and development for a technology that, by design, would reduce its own profit margins, Sánchez-Vizcaíno speculates.....

|
Scooped by
Juan Lama
|
Long before COVID-19, scientists had been working to identify animal viruses that could potentially jump to people. These efforts have led to a Web-based platform called SpillOver, which ranks the risk that various viruses will make the leap. Developers hope the new tool will help public health experts and policymakers avoid future outbreaks. Jonna Mazet, an epidemiologist and disease ecologist at the University of California, Davis, has led this work for more than a decade. It began with the USAID PREDICT project, which sought to go beyond well-tracked influenza viruses and identify other emerging pathogens that pose a risk to humans. Thousands of scientists scoured more than 30 countries to locate and identify animal viruses, discovering many new ones in the process. But not every virus is equally threatening. So Mazet and her colleagues decided to create a framework to interpret their findings. “We wanted to move beyond scientific stamp collecting [simply finding viruses] to actual risk evaluation and reduction,” she says. The team was surprised to find very little existing research on categorizing threats from viruses that are currently found only in animals but are in viral families that can likely cause disease in people. So the researchers started from scratch, identifying 31 factors pertaining to animal viruses (such as how they are transmitted), to their hosts (such as how many and varied they are), and to the environment (human population density, frequency of interaction with hosts, and more). These are summed up in a risk score out of 155; the higher the score, the more likelihood of spillover. Cornell University virologist Colin Parrish, who was not involved in the study, says the factors examined were important in previous spillovers. But he notes that other viruses' crossover risk may be heightened by unforeseeable factors that crop up later. “It's a bit like the stock market,” he says. The new study, published in the Proceedings of the National Academy of Sciences USA, ranks 887 animal-borne viruses. Twelve known human pathogens scored at the top—with the virus that causes COVID-19 in second place, just under the rat-carried Lassa virus. (Influenza would have topped the list if included, Mazet says, but flu variants are already tracked elsewhere.) Parrish notes that the list also omits insect-borne viruses and those from domesticated animals. “This is a work in progress,” he says. “I'm sure it will be iterated into a more powerful tool as more information and data become available. SpillOver is publicly editable, and scientists around the world are already contributing their own findings. Mazet hopes it catches the attention of public health practitioners and leaders, too. With targeted action, Mazet says, “we can ensure that we don't have these spillovers at all. Or if we do, we're ready for them—because we're watching.” See also research published in P.N.A.S. (April 13, 2021): https://doi.org/10.1073/pnas.2002324118

|
Scooped by
Juan Lama
|
Humans are not the only species facing a potential threat from SARS-CoV-2, the novel coronavirus that causes COVID-19, according to a new study from the University of California, Davis. An international team of scientists used genomic analysis to compare the main cellular receptor for the virus in humans -- angiotensin converting enzyme-2, or ACE2 -- in 410 different species of vertebrates, including birds, fish, amphibians, reptiles and mammals. ACE2 is normally found on many different types of cells and tissues, including epithelial cells in the nose, mouth and lungs. In humans, 25 amino acids of the ACE2 protein are important for the virus to bind and gain entry into cells. The researchers used these 25 amino acid sequences of the ACE2 protein, and modeling of its predicted protein structure together with the SARS-CoV-2 spike protein, to evaluate how many of these amino acids are found in the ACE2 protein of the different species. "Animals with all 25 amino acid residues matching the human protein are predicted to be at the highest risk for contracting SARS-CoV-2 via ACE2," said Joana Damas, first author for the paper and a postdoctoral research associate at UC Davis. "The risk is predicted to decrease the more the species' ACE2 binding residues differ from humans." About 40 percent of the species potentially susceptible to SARS-CoV-2 are classified as "threatened" by the International Union for Conservation of Nature and may be especially vulnerable to human-to-animal transmission. The study was published Aug. 21 in the Proceedings of the National Academy of Sciences. "The data provide an important starting point for identifying vulnerable and threatened animal populations at risk of SARS-CoV-2 infection," said Harris Lewin, lead author for the study and a distinguished professor of evolution and ecology at UC Davis. "We hope it inspires practices that protect both animal and human health during the pandemic." Endangered species predicted to be at risk Several critically endangered primate species, such as the Western lowland gorilla, Sumatran orangutan and Northern white-cheeked gibbon, are predicted to be at very high risk of infection by SARS-CoV-2 via their ACE2 receptor. Other animals flagged as high risk include marine mammals such as gray whales and bottlenose dolphins, as well as Chinese hamsters. Domestic animals such as cats, cattle and sheep were found to have a medium risk, and dogs, horses and pigs were found to have low risk for ACE2 binding. How this relates to infection and disease risk needs to be determined by future studies, but for those species that have known infectivity data, the correlation is high. In documented cases of SARS-COV-2 infection in mink, cats, dogs, hamsters, lions and tigers, the virus may be using ACE2 receptors or they may use receptors other than ACE2 to gain access to host cells. Lower propensity for binding could translate to lower propensity for infection, or lower ability for the infection to spread in an animal or between animals once established. Because of the potential for animals to contract the novel coronavirus from humans, and vice versa, institutions including the National Zoo and the San Diego Zoo, which both contributed genomic material to the study, have strengthened programs to protect both animals and humans. "Zoonotic diseases and how to prevent human to animal transmission is not a new challenge to zoos and animal care professionals," said co-author Klaus-Peter Koepfli, senior research scientist at Smithsonian-Mason School of Conservation and former conservation biologist with the Smithsonian Conservation Biology Institute's Center for Species Survival and Center for Conservation Genomics. "This new information allows us to focus our efforts and plan accordingly to keep animals and humans safe." The authors urge caution against overinterpreting the predicted animal risks based on the computational results, noting the actual risks can only be confirmed with additional experimental data. The list of animals can be found here. Research has shown that the immediate ancestor of SARS-CoV-2 likely originated in a species of bat. Bats were found to be at very low risk of contracting the novel coronavirus via their ACE2 receptor, which is consistent with actual experimental data. Whether bats directly transmitted the novel coronavirus directly to humans, or whether it went through an intermediate host, is not yet known, but the study supports the idea that one or more intermediate hosts was involved. The data allow researchers to zero in on which species might have served as an intermediate host in the wild, assisting efforts to control a future outbreak of SARS-CoV-2 infection in human and animal populations. Study published in P.N.A.S. (August 21, 2020):
|



 Your new post is loading...
Your new post is loading...