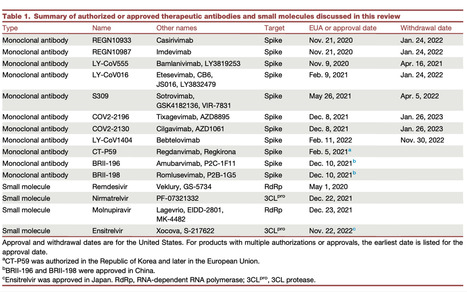
Over four years have passed since the beginning of the COVID-19 pandemic. The scientific response has been rapid and effective, with many therapeutic monoclonal antibodies and small molecules developed for clinical use. However, given the ability for viruses to become resistant to antivirals, it is perhaps no surprise that the field has identified resistance to nearly all of these compounds. Here, we provide a comprehensive review of the resistance profile for each of these therapeutics. We hope that this resource provides an atlas for mutations to be aware of for each agent, particularly as a springboard for considerations for the next generation of antivirals. Finally, we discuss the outlook and thoughts for moving forward in how we continue to manage this, and the next, pandemic.
Published (April 18, 2024):



 Your new post is loading...
Your new post is loading...