Your new post is loading...

|
Scooped by
Juan Lama
|
Hong Kong issued a major health alert as a man battles for his life in intensive care after contracting a deadly form of herpes from monkeys. A concerning health alert has been issued in Hong Kong as a 37-year-old man fights for his life in intensive care after contracting a deadly form of herpes from monkeys. The man's condition has prompted health officials to caution against interactions with wild macaques, urging the public to exercise caution. The victim sustained injuries from wild macaques in February during a visit to Kam Shan Country Park, colloquially known as 'Monkey Hill'. Initially admitted to the hospital with a fever on March 21, his condition deteriorated rapidly, leading to a transfer to the intensive care unit (ICU) where he remains in critical condition. Health authorities confirmed the man's ailment as herpes B virus, also known as herpes simiae, a disease commonly found in macaques. While this virus typically manifests as mild symptoms or remains asymptomatic in monkeys, it poses severe risks to humans, potentially leading to life-threatening complications. Symptoms of the virus in humans can escalate from fever to respiratory issues, nausea, neurological impairments such as numbness or coordination problems, and in severe cases, irreversible brain damage or fatality. Despite being first discovered in 1932, human cases remain rare, with approximately 50 recorded instances and 21 fatalities globally. Dr. Wilson Lam, president of the Hong Kong Society of Infectious Diseases, highlighted the seriousness of the situation saying: "We don’t know the virus very well, but based on the limited data, if humans are in contact with the virus, there’s a high chance of infection. It can have serious health consequences that affect the spinal nerves and central nervous system." The incident marks the first recorded case of human infection with the B virus in Hong Kong, despite the prevalence of macaques in the region. Health officials urged residents and visitors to refrain from touching or feeding wild monkeys to prevent potential transmission of the virus. As investigations continue, authorities stressed the importance of public cooperation in preventing the spread of this potentially lethal virus.

|
Scooped by
Juan Lama
|
The bird flu virus spreading through dairy cattle in the United States may be expanding its reach via milking equipment, the people doing the milking, or both, U.S. Department of Agriculture (USDA) representatives reported today at an international, virtual meeting held to update the situation. The avian virus may not be spreading directly from cows breathing on cows, as some researchers have speculated, according to USDA scientists who took part in the meeting, organized jointly by the World Organisation for Animal Health and the United Nations’s Food and Agricultural Organization. “We haven’t seen any true indication that the cows are actively shedding virus and exposing it directly to other animals,” said USDA’s Mark Lyons, who directs ruminant health for the agency and presented some of its data. The finding might also point to ways to protect humans. So far one worker at a dairy farm with infected cattle was found to have the virus, but no other human cases have been confirmed. USDA researchers tested milk, nasal swabs, and blood from cows at affected dairies and only found clear signals of the virus in the milk. “Right now, we don’t have evidence that the virus is actively replicating within the body of the cow other than the udder,” Suelee Robbe Austerman of USDA’s National Veterinary Services Laboratory told the gathering. The virus might be transmitted from cow to cow in milk droplets on dairy workers’ clothing or gloves, or in the suction cups attached to the udders for milking, Lyons said. (In a 30 March interview with Sciencenone, Thijs Kuiken, a leading avian influenza researcher at Erasmus Medical Center, had suggested the milking machines might be responsible because the components may not always be disinfected between cows.) The influenza virus causing the outbreak, an H5N1 subtype that is known as clade 2.3.4.4b, has devastated wild birds and poultry around the world for more than 2 years, and researchers at first thought migratory birds were responsible for spreading it to all of the affected dairy farms. But USDA scientists now think the movement of cattle, which are frequently transported from the southern parts of the country to the Midwest and north in the spring, may also have played an important role. And they floated the possibility, without naming specific herds or locations, that all affected cows may trace back to a single farm. Since first revealing the infection of dairy cows with this bird flu virus on 25 March, USDA has confirmed it has spread to cattle in six states. And the agency has used the virus’ genetics to track its movements. The virus found on U.S. farms has a specific genetic signature, which led USDA to name it 3.13. “It’s not a common [strain], but it’s very much a descendant of the viruses that have been dominating the flyways in the Pacific and the central flyways in the United States,” Robbe Austerman said. The USDA scientists reported that the agency’s analysis of the different viruses from the cows indicates they likely came from one source. Cows from infected farms in Texas appear to have moved the virus to farms in Idaho, Michigan, and Ohio. “The cow viruses so far have all been similar enough that it would be consistent with a single spillover event or a couple of very closely related spillover events,” Robbe Austerman said. “So far we don’t have any evidence that this is being introduced multiple times into the cows.” The virus in cows could spread to poultry, which has that industry on edge. USDA tightly regulates avian influenza virus strains that are deadly to poultry, requiring culling of entire flocks if one bird tests positive for this H5N1 or one of its relatives; to date, commercial and backyard poultry farms have had to cull 85 million birds because of this virus. But USDA is not calling for any such drastic measures with cow herds. In response to questions from Sciencenone, Ashley Peterson, who handles regulatory affairs for the National Chicken Council, said “out of an abundance of caution, we believe it is prudent to restrict the movement of cows from positive herds.” Although some researchers agree USDA should stop the transport of dairy cows, the agency has so far declined to take that disruptive action. “We heavily rely on the producers who have been isolating the animals within the dairy herds,” Lyons said. USDA also says it has no evidence of beef cattle becoming infected. USDA and the Food and Drug Administration have stressed that pasteurization kills viruses, so there is “no concern” about the safety of commercial milk. They do recommend people not consume raw milk or products made from it. Sources tell Science none some dairy farms that were later shown to be infected first noticed dead cats as early as mid-February. The animals, which often drink spilled milk on farms, “were the canary in the mine,” one said. The cows’ milk was also unusually thick. Those signs, coupled with the discovery of dead birds on the farms, led to the testing of cow milk for the bird flu virus and the 25 March USDA announcement. Oddly, the dead birds on infected farms were not waterfowl, the migratory birds that typically spread the avian flu viruses to poultry, but “peridomestic” species such as grackles, blackbirds, and pigeons. One farm had the virus detected in poultry before it was found in its cows, and although Robbe Austerman said she hesitated to call it the “ancestral strain,” she noted that “all of the detection so far in cattle are clustered around that.” Further studies should clarify how the virus wound up in cows, which sometimes become infected with flu viruses but have never before been shown to have one that causes high mortality in birds. One possibility is that birds infected cows by shedding their droppings in the cows’ feed or water. But bird flu viruses in the past have also spread for many kilometers in the wind, moving from one poultry farm to another. So the current H5N1 strain could have moved from waterfowl to poultry to cattle—or directly from poultry to cattle and then even to the peridomestic birds. “This could be a multifactorial presentation that we’re seeing,” Lyons said. “Lots of questions still to be answered.”

|
Scooped by
Juan Lama
|
BACKGROUND Nirmatrelvir in combination with ritonavir is an antiviral treatment for mild-to-moderate coronavirus disease 2019 (Covid-19). The efficacy of this treatment in patients who are at standard risk for severe Covid-19 or who are fully vaccinated and have at least one risk factor for severe Covid-19 has not been established. METHODS In this phase 2–3 trial, we randomly assigned adults who had confirmed Covid-19 with symptom onset within the past 5 days in a 1:1 ratio to receive nirmatrelvir–ritonavir or placebo every 12 hours for 5 days. Patients who were fully vaccinated against Covid-19 and who had at least one risk factor for severe disease, as well as patients without such risk factors who had never been vaccinated against Covid-19 or had not been vaccinated within the previous year, were eligible for participation. Participants logged the presence and severity of prespecified Covid-19 signs and symptoms daily from day 1 through day 28. The primary end point was the time to sustained alleviation of all targeted Covid-19 signs and symptoms. Covid-19–related hospitalization and death from any cause were also assessed through day 28. RESULTS Among the 1296 participants who underwent randomization and were included in the full analysis population, 1288 received at least one dose of nirmatrelvir–ritonavir (654 participants) or placebo (634 participants) and had at least one postbaseline visit. The median time to sustained alleviation of all targeted signs and symptoms of Covid-19 was 12 days in the nirmatrelvir–ritonavir group and 13 days in the placebo group (P=0.60). Five participants (0.8%) in the nirmatrelvir–ritonavir group and 10 (1.6%) in the placebo group were hospitalized for Covid-19 or died from any cause (difference, −0.8 percentage points; 95% confidence interval, −2.0 to 0.4). The percentages of participants with adverse events were similar in the two groups (25.8% with nirmatrelvir–ritonavir and 24.1% with placebo). In the nirmatrelvir–ritonavir group, the most commonly reported treatment-related adverse events were dysgeusia (in 5.8% of the participants) and diarrhea (in 2.1%). CONCLUSIONS The time to sustained alleviation of all signs and symptoms of Covid-19 did not differ significantly between participants who received nirmatrelvir–ritonavir and those who received placebo. (Supported by Pfizer; EPIC-SR ClinicalTrials.gov number, NCT05011513.) Published in NEJM (April 3, 2024):

|
Scooped by
Juan Lama
|
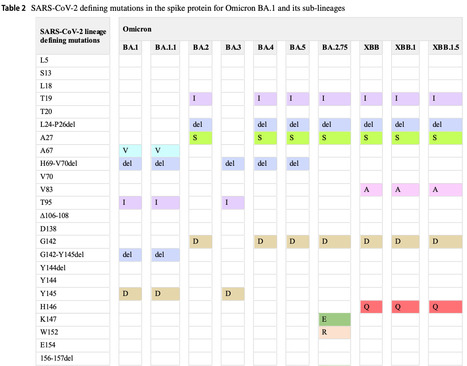
The COVID-19 pandemic has affected hundreds of millions of individuals and caused more than six million deaths. The prolonged pandemic duration and the continual inter-individual transmissibility have contributed to the emergence of a wide variety of SARS-CoV-2 variants. Genomic surveillance and phylogenetic studies have shown that substantial mutations in crucial supersites of spike glycoprotein modulate the binding affinity of the evolved SARS-COV-2 lineages to ACE2 receptors and modify the binding of spike protein with neutralizing antibodies. The immunological spike mutations have been associated with differential transmissibility, infectivity, and therapeutic efficacy of the vaccines and the immunological therapies among the new variants. This review highlights the diverse genetic mutations assimilated in various SARS-CoV-2 variants. The implications of the acquired mutations related to viral transmission, infectivity, and COVID-19 severity are discussed. This review also addresses the effectiveness of human neutralizing antibodies induced by SARS-CoV-2 infection or immunization and the therapeutic antibodies against the ascended variants. Published March 30, 2024: https://doi.org/10.1007/s15010-024-02223-y

|
Scooped by
Juan Lama
|
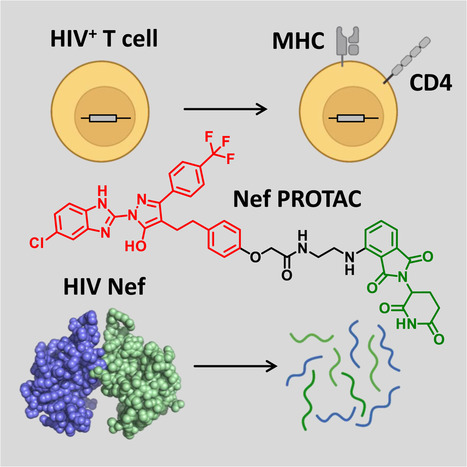
Highlights - HIV-1 Nef enhances the viral life cycle and promotes immune escape of infected cells
- PROTACs promote Nef degradation and form ternary complexes with Nef and cereblon
- PROTACs reverse MHC-I and CD4 downregulation in T cells and block HIV replication
- Targeted degradation may reverse Nef functions to help shrink HIV-1 reservoirs
Summary The HIV-1 Nef accessory factor enhances the viral life cycle in vivo, promotes immune escape of HIV-infected cells, and represents an attractive antiretroviral drug target. However, Nef lacks enzymatic activity and an active site, complicating traditional occupancy-based drug development. Here we describe the development of proteolysis targeting chimeras (PROTACs) for the targeted degradation of Nef. Nef-binding compounds, based on an existing hydroxypyrazole core, were coupled to ligands for ubiquitin E3 ligases via flexible linkers. The resulting bivalent PROTACs induced formation of a ternary complex between Nef and the cereblon E3 ubiquitin ligase thalidomide-binding domain in vitro and triggered Nef degradation in a T cell expression system. Nef-directed PROTACs efficiently rescued Nef-mediated MHC-I and CD4 downregulation in T cells and suppressed HIV-1 replication in donor PBMCs. Targeted degradation is anticipated to reverse all HIV-1 Nef functions and may help restore adaptive immune responses against HIV-1 reservoir cells in vivo. Published March 19, 2024:

|
Scooped by
Juan Lama
|
The Idaho State Department of Agriculture announced that HPAI, known as highly pathogenic avian influenza, or bird flu, has been found in dairy cattle in Idaho. This now brings the number of affected states to four, adding more evidence the virus may be spreading cow to cow. The cows were recently brought into the Cassia County dairy from another state that had found HPAI in dairy cattle, according to the ISDA. The USDA Animal and Plant Health Inspection Service (APHIS) announced that an investigation into mysterious illnesses in dairy cows in three states—Kansas, New Mexico, and Texas—was due to HPAI and that wild birds are the source of the virus. Symptoms of HPAI in cattle include: - Drop in milk production
- Loss of appetite
- Changes in manure consistency
- Thickened or colostrum-like milk
- Low-grade fever
At this stage, there is no concern about the safety of the commercial milk supply or that this circumstance poses a risk to consumer health. The pasteurization process of heating milk to a high temperature ensures milk and dairy products can be consumed safely. The ISDA encourages all dairy producers to closely monitor their herd and contact their local veterinarian immediately if cattle appear to show symptoms. HPAI is a mandatory reportable disease, and any Idaho veterinarians who suspect cases of HPAI in livestock should immediately report it to ISDA at 208-332-8540 or complete the HPAI Livestock Screen at agri.idaho.gov/main/animals/hpai/. APHIS (USDA) update on the dairy cattle bird flu outbreaks (March 29, 2024): https://www.aphis.usda.gov/news/agency-announcements/usda-fda-cdc-share-update-hpai-detections-dairy-cattle

|
Scooped by
Juan Lama
|
Large and giant double-stranded DNA viruses within the phylum Nucleocytoviricota are diverse and prevalent in the environment where they substantially affect the ecology and evolution of eukaryotes. Until now, these viruses were only sporadically found in the digestive system of vertebrates. Here, we present the identification and genomic characterization of a proposed third order of viruses within the class Pokkesviricetes that currently consists of poxviruses and asfuviruses. Members of this newly identified order we provisionally named Egovirales are abundant in the digestive system of vertebrates worldwide and occur in high abundances in >10% of livestock animals, >2% of humans, and wild animals. Egoviruses have linear genomes up to 360 kbp in length that likely produce multilayered icosahedral capsids, similar to those of asfuviruses. The diversity of egoviruses already far exceeds that of all known poxviruses and animal-associated asfuviruses. Phylogenetic analyses and patterns of virus distribution across vertebrates suggest that egoviruses can be either specialists or generalists associated with a single or multiple vertebrate species, respectively. Notably, one egovirus clade is human-specific, evolutionarily constrained, and spread across continents, demonstrating a long-lasting association between Egovirales and the human population on the global scale. Egoviruses not only expand the ecological and evolutionary scope of Pokkesviricetes, but also appear to be the most diverse, widespread, and abundant group of double-stranded DNA viruses infecting eukaryotic cells in the digestive system of vertebrates. Preprint in bioRxiv (March 23, 2024): https://doi.org/10.1101/2024.03.23.586382

|
Scooped by
Juan Lama
|
The Minnesota Board of Animal Health (MBAH) today announced that highly pathogenic avian influenza (HPAI) has been detected in a baby goat that lived on a farm where an outbreak had recently been detected in poultry. Today’s announcement marks the first US detection in livestock. Health officials, including the US Department of Agriculture (USDA), are investigating the transmission of the virus on the farm, which is located in Stevens County in west-central Minnesota. All species on the farm have been placed under quarantine. Poultry had already been quarantined following the February outbreak. "This finding is significant because, while the spring migration is definitely a higher risk transmission period for poultry, it highlights the possibility of the virus infecting other animals on farms with multiple species, " said Minnesota state veterinarian Brian Hoefs, DVM. " Thankfully, research to-date has shown mammals appear to be dead-end hosts, which means they’re unlikely to spread HPAI further...."

|
Scooped by
Juan Lama
|
Immunocompromised people get a new drug to protect against Covid-19 for the first time since the Omicron variants forced another drug off the market. The Food and Drug Administration on Friday authorized a new antibody to protect immunocompromised individuals against Covid-19. The drug, known as Pemgarda and marketed by the biotech Invivyd, is the first such drug to become available since the agency pulled AstraZeneca’s Evusheld off the market in January 2023. New Omicron variants had rendered Evusheld ineffective. Some immunocompromised patients — a group that includes certain cancer patients, patients with some autoimmune or genetic disorders, and organ transplant recipients — have been eagerly awaiting the new prophylactic. Because their condition or drugs they take for it weaken their immune systems, they don’t always get adequate protection from vaccination. Antibody treatments can add additional protection. There remains a need for additional Covid protections. Last week, over 10,000 patients with Covid-19 were admitted to U.S. hospitals. Still, it’s not clear how many patients will avail themselves of the new treatment. Invivyd, the drug’s developer, estimated there are 9 million immunocompromised people in the U.S., but its initial focus will be on the 485,000 with the most acute need: Stem cell transplant recipients, organ transplant recipients, and blood cancer patients. Evusheld itself had gone vastly underused when it was available, prompting the federal government to launch efforts to educate the public about its existence. And the need is probably less acute now for some: Although immunocompromised patients have a reduced response to vaccination, many may still gain some protection from multiple rounds of vaccination and exposure to the virus. Uptake for other measures to mitigate Covid risk, such as boosters and Paxlovid upon infection, has also declined precipitously, despite public health efforts. “We will explore having it available at our institution, so that our very compromised patients and the services and specialities that take care of those patients know that it’s an option,” said Priya Nori, an infectious disease specialist at Albert Einstein College of Medicine. “But we’re not expecting gangbusters in terms of interest.” Invivyd has not yet disclosed a price. The company was originally founded in 2020 by antibody expert Tillman Gerngross, under the name Adagio, and raised $309 million in an IPO a year later. But the company’s original antibody — which had been designed to be all but variant-proof — was soon rendered ineffective by the advent of Omicron, eventually leading to Gerngross’s resignation and a series of high-level changes at the company. Pemgarda is a re-engineered version of the original antibody, designed to cover Omicron variants. That gave Invivyd a faster path to getting a new preventative on the market than competitors who started from scratch. FDA EUA letter (March 22, 2024): https://www.fda.gov/media/177068/download Invivyd Press release: https://investors.invivyd.com/news-releases/news-release-details/invivyd-announces-fda-authorization-emergency-use-pemgardatm

|
Scooped by
Juan Lama
|
Machine-learning system trained on millions of human audio clips shows promise for detecting COVID-19 and tuberculosis. A team led by Google scientists has developed a machine-learning tool that can detect and monitor health conditions by evaluating noises such as coughing and breathing. The artificial intelligence (AI) system, trained on millions of audio clips of human sounds, might one day be used by physicians to diagnose diseases including COVID-19 and tuberculosis and to assess how well a person’s lungs are functioning. This is not the first time a research group has explored using sound as a biomarker for disease. The concept gained traction during the COVID-19 pandemic, when scientists discovered that it was possible to detect the respiratory disease through a person’s cough. What’s new about the Google system — called Health Acoustic Representations (HeAR) — is the massive data set that it was trained on, and the fact that it can be fine-tuned to perform multiple tasks. The researchers, who reported the tool earlier this month in a preprint1 that has not yet been peer reviewed, say it’s too early to tell whether HeAR will become a commercial product. For now, the plan is to give interested researchers access to the model so that they can use it in their own investigations. “Our goal as part of Google Research is to spur innovation in this nascent field,” says Sujay Kakarmath, a product manager at Google in New York City who worked on the project.... Preprint available here https://arxiv.org/pdf/2403.02522.pdf

|
Scooped by
Juan Lama
|
Mammalian cells outpace bacteriophages in the microbial food chain by devouring phages to fuel their growth. The human gut is a bustling highway for a highly diverse microbial community, including an abundance of bacteriophages that modulate the gut microbiome. It’s a phage-infect-bacteria world, and while bacteriophages cannot infect mammalian cells, their paths still intersect. Mammalian cells can engulf phages within the gut. Researchers have observed that different bacteriophages induce opposing reactions such as anti- or proinflammatory responses in mammalian cells. However, it is unclear how bacteriophages interact with cells and modulate these cellular and immune responses. Jeremy Barr, a bacteriophage biologist at Monash University, and his team set out to clarify whether or not phages activate inflammatory pathways. Their findings, published in PLOS Biology, demonstrated that mammalian cells engulfed bacteriophages to fuel cellular growth without inducing inflammation....

|
Scooped by
Juan Lama
|
Highlights - XBB.1.5 COVID-19 mRNA vaccine elicits neutralizing antibodies against current variants
- Depletion of Wuhan-Hu-1 S-reactive plasma antibodies abrogate XBB.1.5 neutralization
- XBB.1.5 COVID-19 mRNA vaccine primary recalls Wuhan-Hu-1 S-reactive memory B cells
Summary Immune imprinting describes how the first exposure to a virus shapes immunological outcomes of subsequent exposures to antigenically related strains. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) Omicron breakthrough infections and bivalent COVID-19 vaccination primarily recall cross-reactive memory B cells induced by prior Wuhan-Hu-1 spike mRNA vaccination rather than priming Omicron-specific naive B cells. These findings indicate that immune imprinting occurs after repeated Wuhan-Hu-1 spike exposures, but whether it can be overcome remains unclear. To understand the persistence of immune imprinting, we investigated memory and plasma antibody responses after administration of the updated XBB.1.5 COVID-19 mRNA vaccine booster. We showed that the XBB.1.5 booster elicited neutralizing antibody responses against current variants that were dominated by recall of pre-existing memory B cells previously induced by the Wuhan-Hu-1 spike. Therefore, immune imprinting persists after multiple exposures to Omicron spikes through vaccination and infection, including post XBB.1.5 booster vaccination, which will need to be considered to guide future vaccination. Published in Immunity (March 14, 2024):

|
Scooped by
Juan Lama
|
Editor’s summary The toolkit for preventing and treating patients with COVID-19 has expanded greatly since the pandemic began. Some of the most effective therapeutics for COVID-19 and, potentially, other coronavirus infections are antivirals that inhibit the SARS-CoV-2 main protease (Mpro). Mpro inhibitors include nirmatrelvir, a component of the oral treatment Paxlovid. However, second-generation drugs are needed, because there is a risk that new SARS-CoV-2 variants could become resistant to nirmatrelvir and other antivirals in clinical use. Here, Westberg et al. used the hepatitis C virus protease inhibitor boceprevir as a starting point to make such a next-generation Mpro inhibitor. Their optimized lead candidate, ML2006a4, exhibited robust antiviral activity in vitro and in mice infected with SARS-CoV-2 and could be administered orally. ML2006a4 appeared less sensitive to mutations in the SARS-CoV-2 Mpro, suggesting that the virus would be less likely to become resistant to ML2006a4 in the real world. —Courtney Malo Abstract Inhibitors of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease (Mpro) such as nirmatrelvir (NTV) and ensitrelvir (ETV) have proven effective in reducing the severity of COVID-19, but the presence of resistance-conferring mutations in sequenced viral genomes raises concerns about future drug resistance. Second-generation oral drugs that retain function against these mutants are thus urgently needed. We hypothesized that the covalent hepatitis C virus protease inhibitor boceprevir (BPV) could serve as the basis for orally bioavailable drugs that inhibit SARS-CoV-2 Mpro more efficiently than existing drugs. Performing structure-guided modifications of BPV, we developed a picomolar-affinity inhibitor, ML2006a4, with antiviral activity, oral pharmacokinetics, and therapeutic efficacy similar or superior to those of NTV. A crucial feature of ML2006a4 is a derivatization of the ketoamide reactive group that improves cell permeability and oral bioavailability. Last, ML2006a4 was found to be less sensitive to several mutations that cause resistance to NTV or ETV and occur in the natural SARS-CoV-2 population. Thus, anticipatory design can preemptively address potential resistance mechanisms to expand future treatment options against coronavirus variants. Published in Science Translational Medicine (March 13, 2024):
|

|
Scooped by
Juan Lama
|
Health Alert Network (HAN). Provided by the Centers for Disease Control and Prevention (CDC). Summary The Centers for Disease Control and Prevention (CDC) is issuing this Health Alert Network (HAN) Health Advisory to inform clinicians, state health departments, and the public of a recently confirmed human infection with highly pathogenic avian influenza (HPAI) A(H5N1) virus in the United States following exposure to presumably infected dairy cattle. The U.S. Department of Agriculture (USDA) recently reported detections of highly pathogenic avian influenza A(H5N1) virus in U.S. dairy cattle in multiple states. This Health Advisory also includes a summary of interim CDC recommendations for preventing, monitoring, and conducting public health investigations of potential human infections with HPAI A(H5N1) virus. Background
A farm worker on a commercial dairy farm in Texas developed conjunctivitis on approximately March 27, 2024, and subsequently tested positive for HPAI A(H5N1) virus infection. HPAI A(H5N1) viruses have been reported in the area’s dairy cattle and wild birds. There have been no previous reports of the spread of HPAI viruses from cows to humans. The patient reported conjunctivitis with no other symptoms, was not hospitalized, and is recovering. The patient was recommended to isolate and received antiviral treatment with oseltamivir. Illness has not been identified in the patient’s household members, who received oseltamivir for post-exposure prophylaxis per CDC Recommendations for Influenza Antiviral Treatment and Chemoprophylaxis. No additional cases of human infection with HPAI A(H5N1) virus associated with the current infections in dairy cattle and birds in the United States, and no human-to-human transmission of HPAI A(H5N1) virus have been identified.
CDC has sequenced the influenza virus genome identified in a specimen collected from the patient and compared it with HPAI A(H5N1) sequences from cattle, wild birds, and poultry. While minor changes were identified in the virus sequence from the patient specimen compared to the viral sequences from cattle, both cattle and human sequences lack changes that would make them better adapted to infect mammals. In addition, there were no markers known to be associated with influenza antiviral drug resistance found in the virus sequences from the patient’s specimen, and the virus is closely related to two existing HPAI A(H5N1) candidate vaccine viruses that are already available to manufacturers, and which could be used to make vaccine if needed. This patient is the second person to test positive for HPAI A(H5N1) virus in the United States. The first case was reported in April 2022 in Colorado in a person who had contact with poultry that was presumed to be infected with HPAI A(H5N1) virus. Currently, HPAI A(H5N1) viruses are circulating among wild birds in the United States, with associated outbreaks among poultry and backyard flocks and sporadic infections in mammals....

|
Scooped by
Juan Lama
|
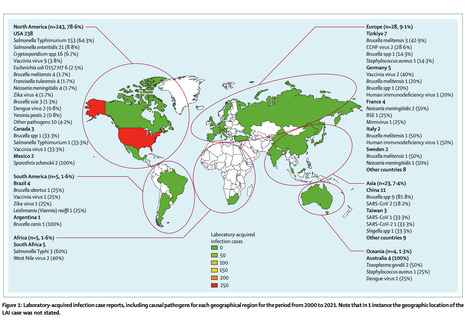
Laboratory-acquired infections (LAIs) and accidental pathogen escape from laboratory settings (APELS) are major concerns for the community. A risk-based approach for pathogen research management within a standard biosafety management framework is recommended but is challenging due to reasons such as inconsistency in risk tolerance and perception. Here, we performed a scoping review using publicly available, peer-reviewed journal and media reports of LAIs and instances of APELS between 2000 and 2021. We identified LAIs in 309 individuals in 94 reports for 51 pathogens. Eight fatalities (2·6% of all LAIs) were caused by infection with Neisseria meningitidis (n=3, 37·5%), Yersinia pestis (n=2, 25%), Salmonella enterica serotype Typhimurium (S Typhimurium; n=1, 12·5%), or Ebola virus (n=1, 12·5%) or were due to bovine spongiform encephalopathy (n=1, 12·5%). The top five LAI pathogens were S Typhimurium (n=154, 49·8%), Salmonella enteritidis (n=21, 6·8%), vaccinia virus (n=13, 4·2%), Brucella spp (n=12, 3·9%), and Brucella melitensis (n=11, 3·6%). 16 APELS were reported, including those for Bacillus anthracis, SARS-CoV, and poliovirus (n=3 each, 18·8%); Brucella spp and foot and mouth disease virus (n=2 each, 12·5%); and variola virus, Burkholderia pseudomallei, and influenza virus H5N1 (n=1 each, 6·3%). Continual improvement in LAI and APELS management via their root cause analysis and thorough investigation of such incidents is essential to prevent future occurrences. The results are biased due to the reliance on publicly available information, which emphasises the need for formalised global LAIs and APELS reporting to better understand the frequency of and circumstances surrounding these incidents. Published in The Lancet Microbre (Feb. 2024): https://doi.org/10.1016/S2666-5247(23)00319-1

|
Scooped by
Juan Lama
|
This is a technical summary of an analysis of the genomic sequences of viruses associated with an outbreak of highly pathogenic avian influenza (HPAI) A(H5N1) viruses in Texas. This analysis supports the conclusion that the overall risk to the general public associated with the ongoing HPAI A(H5N1) outbreak has not changed and remains low at this time. The genome of the virus identified from the patient in Texas is publicly posted in GISAID and has been submitted to GenBank. April 2, 2024 – CDC has sequenced the influenza virus genome identified in a specimen collected from the patient in Texas who was confirmed to be infected with highly pathogenic avian influenza A(H5N1) [“HPAI A(H5N1)”] virus and compared these with HPAI A(H5N1) sequences from cattle, wild birds and poultry. The virus sequences are HA clade 2.3.4.4b HPAI A(H5N1) with each individual gene segment closely related to viruses detected in dairy cattle available from USDA testing in Texas. While minor changes were identified in the virus sequence from the patient specimen compared to the viral sequences from cattle, both cattle and human sequences maintain primarily avian genetic characteristics and for the most part lack changes that would make them better adapted to infect mammals. The genome for the human isolate had one change (PB2 E627K) that is known to be associated with viral adaptation to mammalian hosts, and which has been detected before in people and other mammals infected with HPAI A(H5N1) virus and other avian influenza subtypes (e.g., H7N9), but with no evidence of onward spread among people. Viruses can undergo changes in a host as they replicate after infection. Further, there are no markers known to be associated with influenza antiviral resistance found in the virus sequences from the patient’s specimen and the virus is very closely related to two existing HPAI A(H5N1) candidate vaccine viruses that are already available to manufacturers, and which could be used to make vaccine if needed. Overall, the genetic analysis of HPAI A(H5N1) viruses in Texas supports CDC’s conclusion that the human health risk currently remains low. More details are available in this technical summary below...

|
Scooped by
Juan Lama
|
Health Alert: First Case of Novel Influenza A (H5N1) in Texas, March 2024 Health Alert April 1, 2024 Summary The Texas Department of State Health Services (DSHS) is reporting the first human case of novel avian influenza A(H5N1) in Texas. The patient became ill following contact with dairy cows presumed to be infected with avian influenza. The patient’s primary symptom was conjunctivitis. This is the second case of avian influenza A(H5N1) identified in a person in the United States and is believed to be associated with the recent detections of avian influenza A(H5N1) in dairy cows announced by the Texas Animal Health Commission. DSHS along with local, regional, state, and federal partners, is investigating this ongoing situation. Avian influenza A(H5N1) viruses have only rarely been transmitted from person to person. As such, the risk to the general public is believed to be low; however, people with close contact with affected animals suspected of having avian influenza A(H5N1) have a higher risk of infection. DSHS is issuing this health alert to provide awareness to healthcare providers and ask them to be vigilant for people with signs and symptoms of avian influenza A(H5N1). Suspicion for avian influenza A(H5N1) should be heightened for people who have had contact with animals suspected of having avian influenza A(H5N1)...

|
Scooped by
Juan Lama
|
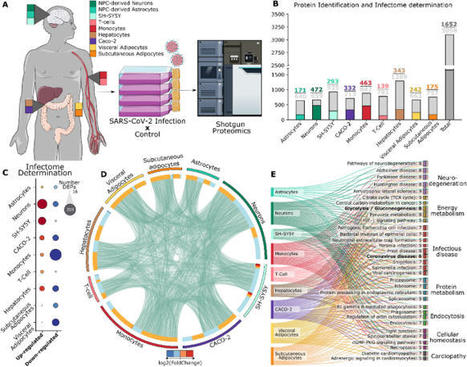
The COVID-19 pandemic was initiated by the rapid spread of a SARS-CoV-2 strain. Though mainly classified as a respiratory disease, SARS-CoV-2 infects multiple tissues throughout the human body, leading to a wide range of symptoms in patients. To better understand how SARS-CoV-2 affects the proteome from cells with different ontologies, this work generated an infectome atlas of 9 cell models, including cells from brain, blood, digestive system, and adipocyte tissue. Our data shows that SARS-CoV-2 infection mainly trigger dysregulations on proteins related to cellular structure and energy metabolism. Despite these pivotal processes, heterogeneity of infection was also observed, highlighting many proteins and pathways uniquely dysregulated in one cell type or ontological group. These data have been made searchable online via a tool that will permit future submissions of proteomic data (https://reisdeoliveira.shinyapps.io/Infectome_App/) to enrich and expand this knowledgebase. Published in Scientific Reports (March 28, 2024): https://doi.org/10.1038/s41598-024-56328-3

|
Scooped by
Juan Lama
|
U.S. regulators confirmed that sick cattle in Texas, Kansas and possibly in New Mexico contracted avian influenza. They stressed that the nation’s milk supply is safe. A highly fatal form of avian influenza, or bird flu, has been confirmed in U.S. cattle in Texas and Kansas, the Department of Agriculture announced on Monday. It is the first time that cows infected with the virus have been identified. The cows appear to have been infected by wild birds, and dead birds were reported on some farms, the agency said. The results were announced after multiple federal and state agencies began investigating reports of sick cows in Texas, Kansas and New Mexico. In several cases, the virus was detected in unpasteurized samples of milk collected from sick cows. Pasteurization should inactivate the flu virus, experts said, and officials stressed that the milk supply was safe. “At this stage, there is no concern about the safety of the commercial milk supply or that this circumstance poses a risk to consumer health,” the agency said in a statement. Outside experts agreed. “It has only been found in milk that is grossly abnormal,” said Dr. Jim Lowe, a veterinarian and influenza researcher at the College of Veterinary Medicine at the University of Illinois at Urbana-Champaign. In those cases, the milk was described as thick and syrupy, he said, and was discarded. The agency said that dairies are required to divert or destroy milk from sick animals.... USDA report (March 25, 2024): https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2024/hpai-cattle

|
Scooped by
Juan Lama
|
Persistent SARS-CoV-2 infections may act as viral reservoirs that could seed future outbreaks, give rise to highly divergent lineages and contribute to cases with post-acute COVID-19 sequelae (long COVID). However, the population prevalence of persistent infections, their viral load kinetics and evolutionary dynamics over the course of infections remain largely unknown. Here, using viral sequence data collected as part of a national infection survey, we identified 381 individuals with SARS-CoV-2 RNA at high titre persisting for at least 30 days, of which 54 had viral RNA persisting at least 60 days. We refer to these as ‘persistent infections’ as available evidence suggests that they represent ongoing viral replication, although the persistence of non-replicating RNA cannot be ruled out in all. Individuals with persistent infection had more than 50% higher odds of self-reporting long COVID than individuals with non-persistent infection. We estimate that 0.1–0.5% of infections may become persistent with typically rebounding high viral loads and last for at least 60 days. In some individuals, we identified many viral amino acid substitutions, indicating periods of strong positive selection, whereas others had no consensus change in the sequences for prolonged periods, consistent with weak selection. Substitutions included mutations that are lineage defining for SARS-CoV-2 variants, at target sites for monoclonal antibodies and/or are commonly found in immunocompromised people. This work has profound implications for understanding and characterizing SARS-CoV-2 infection, epidemiology and evolution. Using viral sequence data, individuals with persistent SARS-CoV-2 infections were identified, and had higher odds of self-reporting long COVID, in a large community surveillance study. Published in Nature (Feb. 21, 2024): https://doi.org/10.1038/s41586-024-07029-4

|
Scooped by
Juan Lama
|
Introduction Continued SARS-CoV-2 infection among immunocompromised individuals is likely to play a role in generating genomic diversity and the emergence of novel variants. Antiviral treatments such as molnupiravir are used to mitigate severe COVID-19 outcomes, but the extended effects of these drugs on viral evolution in patients with chronic infections remain uncertain. This study investigates how molnupiravir affects SARS-CoV-2 evolution in immunocompromised patients with prolonged infections. Methods The study included five immunocompromised patients treated with molnupiravir and four patients not treated with molnupiravir (two immunocompromised and two non-immunocompromised). We selected patients who had been infected by similar SARS-CoV-2 variants and with high-quality genomes across timepoints to allow comparison between groups. Throat and nasopharyngeal samples were collected in patients up to 44 days post treatment and were sequenced using tiled amplicon sequencing followed by variant calling. The UShER pipeline and University of California Santa Cruz genome viewer provided insights into the global context of variants. Treated and untreated patients were compared, and mutation profiles were visualised to understand the impact of molnupiravir on viral evolution. Findings Patients treated with molnupiravir showed a large increase in low-to-mid-frequency variants in as little as 10 days after treatment, whereas no such change was observed in untreated patients. Some of these variants became fixed in the viral population, including non-synonymous mutations in the spike protein. The variants were distributed across the genome and included unique mutations not commonly found in global omicron genomes. Notably, G-to-A and C-to-T mutations dominated the mutational profile of treated patients, persisting up to 44 days post treatment. Interpretation Molnupiravir treatment in immunocompromised patients led to the accumulation of a distinctive pattern of mutations beyond the recommended 5 days of treatment. Treated patients maintained persistent PCR positivity for the duration of monitoring, indicating clear potential for transmission and subsequent emergence of novel variants. Published in The Lancet Microbe (March 22, 2024):

|
Scooped by
Juan Lama
|
The SARS-CoV-2 variant JN.1 swiftly became the global dominant strain due to a spike protein Leu455Ser substitution, boosting transmissibility and immune-escape capabilities, surpassing its predecessor BA.2.86 and other variants. These alterations have resulted in a surge of COVID-19 cases, reflected in wastewater-surveillance data surpassing rates, observed during the initial omicron wave. However, concerns persist that JN.1 might have an increased capacity to replicate in the gut, potentially leading to infected individuals shedding a higher number of viral copies than previously seen. As there is currently a lack of available data for fecal viral shedding, we are presenting the initial longitudinal and quantitative faecal shedding data for SARS-CoV-2 RNA in individuals infected with XBB.1.5, EG.5.1, HV.1, JD.1.1, BA.2.86, and JN.1. 856 faecal samples were obtained from 113 non-hospitalised individuals with confirmed PCR positivity for SARS-CoV-2 RNA. Variants were identified through Sanger sequencing. Detailed protocols for processing and extracting SARS-CoV-2 RNA from stool samples are provided in the appendix... Published in The Lancet Infectious Diseases (March 21, 2024):

|
Scooped by
Juan Lama
|
Significance The Achilles heel of the best available antiviral mRNA vaccines and biomolecular drugs includes the loss of their potency due to viral mutations and thermal exposure. Chiral L- or D-penicillamine-coated CuS nanoparticles (NPs) provide an additional pathway to address these problems. The geometric complementary of their twisted conical shape to the spike protein of SARS-CoV-2 results in agglutination of the virus with antibody-like efficiency. Strong nanoparticle–protein interactions lead to broad-spectrum antiviral activity for several SARS-CoV-2 variants. An inhalable nano-formulation effectively protected mice from SARS-CoV-2 infection for 72 h. The combination of temperature robustness, curative capabilities, and broad activity makes possible utilization of chiral NPs as rapid-deployment antivirals for first responders and emergency biomedical stockpiles for potential pandemics. Abstract The incessant mutations of viruses, variable immune responses, and likely emergence of new viral threats necessitate multiple approaches to novel antiviral therapeutics. Furthermore, the new antiviral agents should have broad-spectrum activity and be environmentally stable. Here, we show that biocompatible tapered CuS nanoparticles (NPs) efficiently agglutinate coronaviruses with binding affinity dependent on the chirality of surface ligands and particle shape. L-penicillamine-stabilized NPs with left-handed curved apexes display half-maximal inhibitory concentrations (IC50) as low as 0.66 pM (1.4 ng/mL) and 0.57 pM (1.2 ng/mL) for pseudo-type SARS-CoV-2 viruses and wild-type Wuhan-1 SARS-CoV-2 viruses, respectively, which are about 1,100 times lower than those for antibodies (0.73 nM). Benefiting from strong NPs–protein interactions, the same particles are also effective against other strains of coronaviruses, such as HCoV-HKU1, HCoV-OC43, HCoV-NL63, and SARS-CoV-2 Omicron variants with IC50 values below 10 pM (21.8 ng/mL). Considering rapid response to outbreaks, exposure to elevated temperatures causes no change in the antiviral activity of NPs while antibodies are completely deactivated. Testing in mice indicates that the chirality-optimized NPs can serve as thermally stable analogs of antiviral biologics complementing the current spectrum of treatments. Published in PNAS (Jan. 19, 2024):

|
Scooped by
Juan Lama
|
Filoviruses frequently emerge to cause terrifying outbreaks of often-fatal human disease. Treatment options so far have focused on monoclonal antibodies. Remdesivir is an adenosine analog that binds viral RNA polymerase to block replication by premature termination of RNA transcription. The drug has been successfully used intravenously for treating progressive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in humans. Cross et al. tested a related oral prodrug, obeldesivir (currently in phase 3 clinical trials for COVID-19 treatment), in nonhuman primates for its therapeutic value against filoviruses (see the Perspective by Sprecher and Van Herp). When administered to the animals within 24 hours of virus exposure once daily for 10 days, the drug conferred complete protection against lethal infection with Sudan ebolavirus. Having an oral drug would be a major logistical advantage for use in the remote, resource-poor areas where filoviruses occur. —Caroline Ash Published in Science (March 15, 2024): https://doi.org/10.1126/science.adk6176

|
Scooped by
Juan Lama
|
Research shows that even mild COVID-19 can lead to the equivalent of seven years of brain aging. From the very early days of the pandemic, brain fog emerged as a significant health condition that many experience after COVID-19. Brain fog is a colloquial term that describes a state of mental sluggishness or lack of clarity and haziness that makes it difficult to concentrate, remember things and think clearly. Fast-forward four years and there is now abundant evidence that being infected with SARS-CoV-2 – the virus that causes COVID-19 – can affect brain health in many ways. In addition to brain fog, COVID-19 can lead to an array of problems, including headaches, seizure disorders, strokes, sleep problems, and tingling and paralysis of the nerves, as well as several mental health disorders. A large and growing body of evidence amassed throughout the pandemic details the many ways that COVID-19 leaves an indelible mark on the brain. But the specific pathways by which the virus does so are still being elucidated, and curative treatments are nonexistent. Now, two new studies published in the New England Journal of Medicine shed further light on the profound toll of COVID-19 on cognitive health.....
|



 Your new post is loading...
Your new post is loading...