Researchers developed a new detection assay for SARS-CoV-2.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|

Loïc Lepiniec's curator insight,
March 24, 2016 2:53 AM
In this work, the authors adapt self-complementing split fluorescent proteins as epitope tags for live cell protein labelling. The two tags, GFP11 and sfCherry11 are derived from the eleventh β-strand of super-folder GFP and sfCherry, respectively. The small size of FP11-tags enables a cost-effective and scalable way to insert them into endogenous genomic loci via CRISPR-mediated homology-directed repair. Their results demontrate the versatility of FP11-tag as a labelling tool as well as a multimerization-control tool for both imaging and non-imaging applications. |




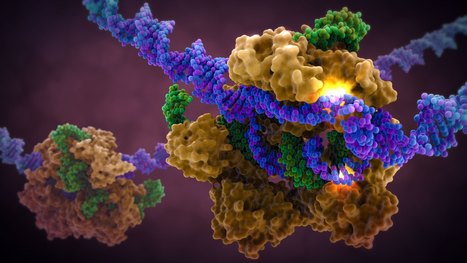

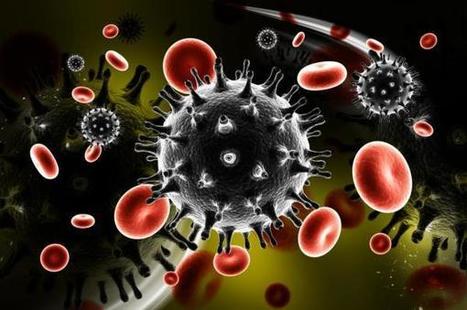








In a recent study published in Proceedings of the National Academy of Sciences, researchers developed a new assay for the detection of SARS-CoV-2. For the test, CRISPR systems were coupled to loop-mediated isothermal amplification (LAMP) or recombinase polymerase (RPA) amplification to boost sensitivity and improve detection at attomolar levels. In addition, reverse transcription (RT)-RPA/LAMP systems coupled with Cas13 systems have been developed for the detection of SARS-CoV-2. RT-LAMP has become the preferred method due to its high sensitivity, low cost and ease of use. Since these systems use functional Cas enzymes at 37°C and LAMP requires high temperatures (55°C to 65°C), the RT-LAMP/RPA assays coupled with CRISPR. The multiplexed assay therefore has a thermophilic Cas13a isolated from Thermoclostridium caenicola (TccCas13a) and uses it for the detection of SARS-CoV-2 using fluorescent amidite-labelled RNA and has developed an AapCas12b-based RNase P detection using HEX-labelled ssDNA reporters. The researchers achieved simultaneous detection of RNase P and SARS-CoV-2 without any interference from the fluorescence signals. A mobile phone application was developed to collect and interpret the fluorescence viewer readings at low cost.