For the first time, scientists have created from scratch self-assembling protein filaments built from identical protein subunits that snap together spontaneously to form long, helical, thread-like structures.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|







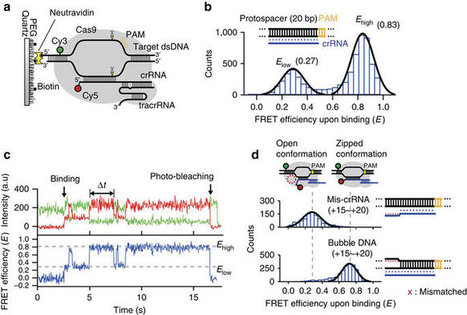


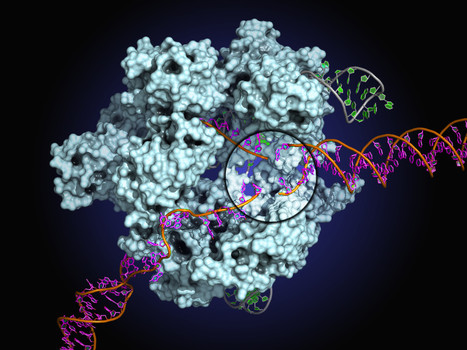


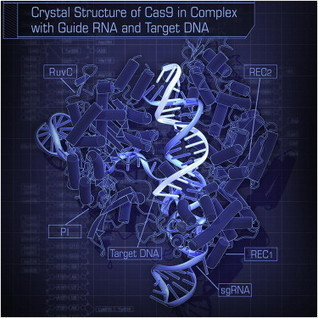







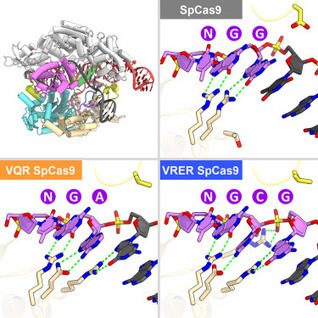










This protein-design advance could lead to creatiing materials unlike any in nature and with a range of applications, from diagnostics to nano-electronics.
Published in Science
De novo design of self-assembling helical protein filaments