We have been developing CRISPR-directed gene editing as an augmentative therapy for the treatment of non-small cell lung carcinoma (NSCLC) by genetic disruption of Nuclear Factor Erythroid 2-Related Factor 2 (NRF2). NRF2 promotes tumor cell survival in response to therapeutic intervention and thus its disablement should restore or enhance effective drug action. Here, we report how NRF2 disruption leads to collateral damage in the form of CRISPR-mediated exon skipping. Heterogeneous populations of transcripts and truncated proteins produce a variable response to chemotherapy, dependent on which functional domain is missing. We identify and characterize predicted and unpredicted transcript populations and discover that several types of transcripts arise through exon skipping; wherein one or two NRF2 exons are missing. In one specific case, the presence or absence of a single nucleotide determines whether an exon is skipped or not by reorganizing Exonic Splicing Enhancers (ESEs). We isolate and characterize the diversity of clones induced by CRISPR activity in a NSCLC tumor cell population, a critical and often overlooked genetic byproduct of this exciting technology. Finally, gRNAs must be designed with care to avoid altering gene expression patterns that can account for variable responses to solid tumor therapy.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|






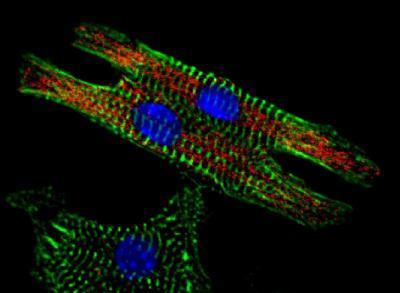








There is a pervasive sense of hopelessness in these non-small cell lung cancer (NSCLC) patients due to the current lack of treatment options, treatment failure due to drug resistance, and poor survival rates. Chemotherapy resistance is a major challenge in the treatment of NSCLC. Nuclear factor erythroid 2-related factor 2 (NRF2) may be at the root of this resistance. It is a master regulator of hundreds of other genes involved in numerous cytoprotective and metabolic pathways. Under normal physiological conditions, NRF2 protects cells from oxidative stress, toxic attacks and chemotherapy, keeping cells in homeostasis. Cancer cells hijack the NRF2 pathway and disrupt this function. When NRF2 becomes over-expressed and accumulates in the cell, the cell is able to fight off these toxic insults. This is really where the cancer becomes chemoresistant. Advanced patients have much higher levels of NRF2. However, in a paper published last month in Gene Therapy, a research team describes a new anti-cancer strategy that uses CRISPR-Cas9 to eliminate NRF2. The key finding of the study is that CRISPR-based disruption of NRF2 can lead to exon skipping. A dual sgRNA approach to target exon 4 resulted in the deletion of a 103-base-pair fragment, including exonic splice enhancer sequences.