RAG2-SCID is a primary immunodeficiency caused by mutations in Recombination-activating gene 2 (RAG2), a gene intimately involved in the process of lymphocyte maturation and function. ex-vivo manipulation of a patient’s own hematopoietic stem and progenitor cells (HSPCs) using CRISPR-Cas9/rAAV6 gene editing could provide a therapeutic alternative to the only current treatment, allogeneic hematopoietic stem cell transplantation (HSCT). Here we show an innovative RAG2 correction strategy that replaces the entire endogenous coding sequence (CDS) for the purpose of preserving the critical endogenous spatiotemporal gene regulation and locus architecture. Expression of the corrective transgene leads to successful development into CD3+TCRαβ+ and CD3+TCRγδ+ T cells and promotes the establishment of highly diverse TRB and TRG repertoires in an in-vitro T-cell differentiation platform. Thus, our proof-of-concept study holds promise for safer gene therapy techniques of tightly regulated genes. RAG2-SCID is a primary immunodeficiency caused by mutations in Recombination-activating gene 2 (RAG2). Here the authors report a RAG2 correction strategy that replaces the entire endogenous coding sequence (CDS) to maintain the endogenous spatiotemporal gene regulation and locus architecture.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account

| Tags |
|---|
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
Rochester Institute of Technology researchers are improving non-invasive treatment options for degenerative disc disease, an ailment that impacts 3 million adults yearly in the U.S., according to the Mayo Clinic.
BigField GEG Tech's insight:
Researchers are improving non-invasive treatment options for degenerative disc disease, a condition that affects 3 million adults each year in the U.S. Stem cell therapy has been a viable field in regenerative technologies for many pathologies for several years. However, the degenerated intervertebral disc provides a hostile environment that is detrimental to stem cell survival, resulting in limited clinical success of stem cell therapy for the disc. Previous research has shown that stem cell-derived electrical vehicles contain many therapeutically beneficial proteins, lipids and nucleic acids and carry much of the regenerative potential of stem cells. However, by using CRISPR in mesenchymal stem cells, researchers have added to the growing field of regenerative medicine the process of producing cell-based therapies to alleviate pain and lack of mobility. In their study, the researchers target TSG6, an essential stem cell marker known to be linked to the regenerative and anti-inflammatory properties of these stem cells. Their hypothesis is that if they CRISPR-activate TSG6 in stem cells, they will not only increase TSG6 protein levels in the extracellular vesicle cargo, but potentially amplify the stem cells' anti-inflammatory and regenerative properties.
Scientists at Leipzig University, in collaboration with colleagues at Vilnius University in Lithuania, have developed a new method to measure the smallest twists and torques of molecules within milliseconds.
BigField GEG Tech's insight:
With the help of an embedded RNA, CRISPR complexes recognize a short DNA sequence. To better understand this process, researchers took advantage of the fact that the DNA double helix of the target sequence is unwound during recognition to allow base pairing with the RNA. Using a technique known as DNA origami, the researchers constructed a 75 nm-long DNA rotor arm with a gold nanoparticle attached to its end. In the experiment, the unwinding of the 2 nm thick, 10 nm long DNA sequence was transferred to the rotation of the gold nanoparticle along a 160 nm diameter circle: this movement could be amplified and tracked using a special microscope setup. With this new method, the researchers were able to observe sequence recognition by the CRISPR complex almost base pair by base pair. Surprisingly, base pairing with RNA is not energetically advantageous, meaning that the complex is only unstably bound during sequence recognition. Only when the entire sequence is recognized does stable binding occur, and the DNA is then destroyed. The sometimes erroneous results are due to its stochastic nature, i.e. random molecular movements.
Vaccine boosting modifies CAR T cell metabolism and promotes crosstalk between CAR
BigField GEG Tech's insight:
Engineering T cells to destroy cancer cells has proved effective in treating certain types of cancer. However, it has not worked as well for solid tumors. One of the reasons for this lack of success is that T cells only target a single antigen. If some of the tumor cells don't express this antigen, they can evade T cell attack. In 2019, researchers found a way to overcome this obstacle, using a vaccine that enhances CAR-T cell efficacy. In a recent study, researchers wanted to investigate how this additional T-cell response is activated. The researchers discovered that in these vaccinated mice, metabolic changes occur in CAR-T cells that increase their production of interferon gamma, a cytokine that helps stimulate a strong immune response. This helps T cells overcome the tumor's immunosuppressive environment, which normally shuts down all T cells in the vicinity. As CAR-T cells killed tumor cells expressing the target antigen, host T cells encountered other antigens from these tumor cells, stimulating these host T cells to target these antigens and help destroy the tumor cells. Without this host T-cell response, the researchers found that tumors would regrow even if CAR-T cells destroyed most of them.
Molecular routes to metastatic dissemination are critical determinants of aggressive cancers. Through in vivo CRISPR–Cas9 genome editing, we generated somatic mosaic genetically engineered models that faithfully recapitulate metastatic renal tumors. Disruption of 9p21 locus is an evolutionary driver to systemic disease through the rapid acquisition of complex karyotypes in cancer cells. Cross-species analysis revealed that recurrent patterns of copy number variations, including 21q loss and dysregulation of the interferon pathway, are major drivers of metastatic potential. In vitro and in vivo genomic engineering, leveraging loss-of-function studies, along with a model of partial trisomy of chromosome 21q, demonstrated a dosage-dependent effect of the interferon receptor genes cluster as an adaptive mechanism to deleterious chromosomal instability in metastatic progression. This work provides critical knowledge on drivers of renal cell carcinoma progression and defines the primary role of interferon signaling in constraining the propagation of aneuploid clones in cancer evolution. Using genetically engineered models, Genovese and colleagues study patterns of convergent evolution in renal cancer, and pinpoint dysregulation of interferon signaling as a means of adaptation to chromosomal instability in metastatic progression.
BigField GEG Tech's insight:
One of the hallmarks of renal cell carcinoma (RCC) is chromosomal instability, which is associated with resistance to many types of therapy and a poor prognosis. It is not known whether specific types of chromosomal abnormalities are involved in metastasis and how tumors are able to tolerate them. Researchers have used CRISPR/Cas9-based genome editing to generate models of aggressive renal cell carcinoma (RCC), lacking common tumor suppressor genes. Using genome sequencing and single-cell RNA sequencing to examine these models in more detail, the researchers uncovered the molecular drivers of RCC and gained a new understanding of the evolution of chromosomal instability. IFNR gene clusters are normally involved in the immune response. After analyzing various datasets from mice and humans, the researchers discovered an inverse correlation between the loss of these IFNR genes and aneuploidy, a condition characterized by an abnormal number of chromosomes. This study suggests that tumors adapt to high levels of chromosomal instability by disrupting the IFNR pathway, and that this is likely to be a major biomarker of metastatic potential. In future, the researchers plan to test drug combinations in these newly generated models to determine how tumors adapt to various therapies.
Three young patients with relapsed T-cell leukemia have now been treated with base-edited T-cells, as part of a 'bench-to-bedside' collaboration between UCL and Great Ormond Street Hospital for Children (GOSH).
BigField GEG Tech's insight:
Three young patients with relapsing T-cell leukemia have now been treated with CAR T cells. Data from the clinical trial show how the donor CAR T cells were engineered using cutting-edge gene-editing technology to modify single letters of their DNA code so that they could fight leukemia. To generate "universal" anti-T CAR T cell banks for the study, the researchers used T cells from healthy donors. They then made modifications to the cells using basic editing. The steps were: the deletion of existing receptors so that a donor's T cells could be banked and used without pairing, making them "universal", the deletion of a "flag" called CD7 so that T cells would not kill each other, the deletion of a second "flag" called CD52 to make the edited cells invisible to some of the potent drugs administered to the patient during the treatment process, and the addition of a chimeric antigen receptor that recognizes the CD7 T-cell receptor on leukemic T-cells to combat T-cell leukemia. The clinical trial for this treatment is still open and aims to recruit up to 10 patients. The teams hope that the treatment can be offered earlier in the treatment pathway. With additional funding, they also hope to make it available for adults in the future.
The most common form of genetic heart disease is hypertrophic cardiomyopathy (HCM), which is caused by variants in cardiac sarcomeric genes and leads to abnormal heart muscle thickening. Complications of HCM include heart failure, arrhythmia and sudden cardiac death. The dominant-negative c.1208G>A (p.R403Q) pathogenic variant (PV) in β-myosin (MYH7) is a common and well-studied PV that leads to increased cardiac contractility and HCM onset. In this study we identify an adenine base editor and single-guide RNA system that can efficiently correct this human PV with minimal bystander editing and off-target editing at selected sites. We show that delivery of base editing components rescues pathological manifestations of HCM in induced pluripotent stem cell cardiomyocytes derived from patients with HCM and in a humanized mouse model of HCM. Our findings demonstrate the potential of base editing to treat inherited cardiac diseases and prompt the further development of adenine base editor-based therapies to correct monogenic variants causing cardiac disease. Adenine base editing successfully corrected a MYH7 pathogenic variant that causes hypertrophic cardiomyopathy in human cardiomyocytes and a mouse model of the disease, highlighting the potential of the approach to correct monogenic variants causing cardiac disease.
BigField GEG Tech's insight:
The thickening of the heart muscle associated with hypertrophic cardiomyopathy leads to arrhythmia, heart failure and sudden cardiac death. The disease often goes undiagnosed, and heart transplantation is the only known treatment. Researchers have been working on a therapy using base editing to correct one of the mutations causing HCM. In this latest study, their target was a mutation in the myosin heavy chain 7 (MYH7) gene. MYH7 encodes β-myosin heavy chain, a motor ATPase that incorporates into heart muscle filaments and affects their contractions. Moreover, the pathogenic G-to-A mutation in the MYH7 gene studied is responsible for a severe, early-onset form of Crohn's disease. The team used the CRISPR-Cas 9 system to correct the MYH7 mutation in induced pluripotent stem cells derived from MHC patients, achieving extremely high editing efficiencies of up to 99%, with little or no off-target. The next step will be to obtain more data in a large animal model and make this potential therapy as safe as possible.
Chimeric antigen receptors (CARs) and synthetic Notch (synNotch) receptors are engineered cell-surface receptors that sense a target antigen and respond by activating T cell receptor signaling or a customized gene program, respectively. Here, to expand the targeting capabilities of these receptors, we develop “universal” receptor systems for which receptor specificity can be directed post-translationally via covalent attachment of a co-administered antibody bearing a benzylguanine (BG) motif. A SNAPtag self-labeling enzyme is genetically fused to the receptor and reacts with BG-conjugated antibodies for covalent assembly, programming antigen recognition. We demonstrate that activation of SNAP-CAR and SNAP-synNotch receptors can be successfully targeted by clinically relevant BG-conjugated antibodies, including anti-tumor activity of SNAP-CAR T cells in vivo in a human tumor xenograft mouse model. Finally, we develop a mathematical model to better define the parameters affecting universal receptor signaling. SNAP receptors provide a powerful strategy to post-translationally reprogram the targeting specificity of engineered cells. Chimeric antigen receptors (CARs) and synthetic Notch (synNotch) receptors are promising platforms for cell-based immunotherapies. Here, the authors develop highly programmable versions of these receptors that can be universally targeted to antigens of interest through covalent enzyme chemistry.
BigField GEG Tech's insight:
Researchers have developed a universal receptor system that allows T cells to recognize any cell surface target, enabling highly customizable CAR T cell and other immunotherapies for treating cancer and other diseases. The new approach involves engineering T cells with receptors bearing a universal "SNAPtag" that fuses with antibodies targeting different proteins. By tweaking the type or dose of these antibodies, treatments could be tailored for optimal immune responses. The researchers showed that their SNAP approach works in two important receptors: CAR receptors, a synthetic T cell receptor that coordinates a suite of immune responses, and SynNotch, a synthetic receptor that can be programmed to activate just about any gene. In a mouse model of cancer, treatment with SNAP-CAR T cells shrunk tumors and greatly prolonged survival, an important proof-of-concept that sets the stage to test this approach in clinical trials in partnership with Coeptis Therapeutics, which has licensed the SNAP-CAR technology from Pitt. The discovery could extend into solid tumors and give more patients access to the game-changing results CAR T cell therapy has produced in certain blood cancers. With the addition of SNAP, the possibilities for customized therapies become almost endless.
BigField GEG Tech's insight:
A phase I clinical trial of an adjuvant personalized mRNA neoantigen vaccine, autogene cevumeran, in patients with pancreatic ductal carcinoma demonstrates that the vaccine can induce T cell activity that may correlate with delayed recurrence of disease.
Adoptive transfer of genetically engineered chimeric antigen receptor (CAR) T cells is becoming a promising treatment option for hematological malignancies. However, T cell immunotherapies have mostly failed in individuals with solid tumors. Here, with a CRISPR–Cas9 pooled library, we performed an in vivo targeted loss-of-function screen and identified ST3 β-galactoside α-2,3-sialyltransferase 1 (ST3GAL1) as a negative regulator of the cancer-specific migration of CAR T cells. Analysis of glycosylated proteins revealed that CD18 is a major effector of ST3GAL1 in activated CD8+ T cells. ST3GAL1-mediated glycosylation induces the spontaneous nonspecific tissue sequestration of T cells by altering lymphocyte function-associated antigen-1 (LFA-1) endocytic recycling. Engineered CAR T cells with enhanced expression of βII-spectrin, a central LFA-1-associated cytoskeleton molecule, reversed ST3GAL1-mediated nonspecific T cell migration and reduced tumor growth in mice by improving tumor-specific homing of CAR T cells. These findings identify the ST3GAL1–βII-spectrin axis as a major cell-intrinsic program for cancer-targeting CAR T cell migration and as a promising strategy for effective T cell immunotherapy. CAR T cell success requires targeting tumors, but these cells can get trapped in other tissues, such as in the lungs, where they can cause pathology. Here, the authors use a loss-of-function CRISPR screen to identify regulators of CAR T cell tumor trafficking and engineer CAR T cells accordingly to overcome this limitation.
BigField GEG Tech's insight:
Immunotherapy, particularly CAR T-Cell cancer therapy, extends the lives of many patients. But sometimes the therapy randomly migrates to places it shouldn't go, sneaking into the lungs or other non-cancerous tissue and causing toxic side effects. However, a team of researchers has discovered the molecule responsible for guiding T cells to tumors, setting the stage for scientists to improve the revolutionary treatment. Their discovery of the crucial migration control gene that expresses ST3GAL1 is the result of "unbiased genomic screening": researchers used a state-of-the-art CRISPR technique to edit thousands of genes expressed in T cells, then tested the migration control capabilities of these genes, one by one over a period of nearly four years, in mouse models. The next step is to find a drug that can manipulate the key T cell protein, ST3GAL1. If the study progresses as planned, such a drug could be added to the CAR T-cell regimen to ensure that the T cells reach their targets
The expanding applications of nonviral genomic medicines in the lung remain restricted by delivery challenges. Here, leveraging a high-throughput platform, we synthesize and screen a combinatorial library of biodegradable ionizable lipids to build inhalable delivery vehicles for messenger RNA and CRISPR–Cas9 gene editors. Lead lipid nanoparticles are amenable for repeated intratracheal dosing and could achieve efficient gene editing in lung epithelium, providing avenues for gene therapy of congenital lung diseases. A high-throughput screen improves lipid nanoparticle delivery of gene editors in the lung.
BigField GEG Tech's insight:
Researchers are developing lipid nanoparticles that may target the lungs. The particles are made of molecules that contain two parts: a positively charged head group and a long lipid tail. The positive charge of the head group helps the particles interact with negatively charged mRNA, and also helps the mRNA escape from cellular structures that engulf the particles once they enter the cells. In tests on mice, the researchers showed that they could use the particles to deliver mRNA encoding CRISPR/Cas9 components designed to turn off a genetically encoded stop signal in the animals' lung cells. When this stop signal is removed, a gene for a fluorescent protein lights up. Measuring this fluorescent signal allows researchers to determine what percentage of cells have successfully expressed the mRNA. After one dose of mRNA, about 40% of lung epithelial cells were transfected, the researchers found. Two doses brought the level to more than 50% and three doses to 60%. The most important targets for treating lung disease are two types of epithelial cells called club cells and hair cells, and each was transfected at about 15%. These particles could offer an inhalable treatment for cystic fibrosis and other lung diseases.
Researchers from Utah State University and Germany’s Helmholtz Institute for RNA-based Infection Research demonstrate that upon recognition of an RNA target, Cas12a2 cleaves all the other nucleic acids present, destroying the bacterial cell and preventing bacteriophage from replicating.
BigField GEG Tech's insight:
Despite its phylogenetic placement with DNA-targeting nucleases, Cas12a2 targets and cleaves RNA. When researchers put Cas12a2 in test tubes with pure DNA and a guide, nothing happened, because its target was not present and it remained inactive. However, sometimes a small amount of activating RNA was present, contaminating the sample. When the RNA was present and Cas12a2 recognized its target, it destroyed all nucleic acids in the tube. Unlike the CRISPR-Cas9 system, when Cas12a2 finds its target, the infected cell(s) die. This mechanism is known as abortive infection. One of the obvious applications of Cas12a2 is in CRISPR diagnostics. It can easily be reprogrammed to detect certain targets, such as RNA viruses, with high specificity. The researchers have already demonstrated the feasibility of this approach in their recent work. Cas12a2 could also be programmed to kill specific cell types, such as tumor cells, for therapeutic purposes.
The genetic mutation underlying a severe immune system deficiency can be corrected
BigField GEG Tech's insight:
The rare and fatal genetic disease CD3 delta severe combined immunodeficiency, also known as CD3 delta SCID, is caused by a mutation in the CD3D gene, which prevents the production of the CD3 delta protein needed for normal T cell development from blood stem cells. Currently, bone marrow transplantation is the only treatment available, but the procedure carries significant risks. In a study published in Cell, researchers showed that a new genome editing technique called base editing can correct the mutation that causes CD3 delta SCID in blood stem cells and restore their ability to produce T cells. The basic editor corrected an average of nearly 71 percent of the patient's stem cells in three experiments. The researchers then tested whether the corrected cells could give rise to T cells. When the corrected blood stem cells were introduced into artificial thymic organoids, they produced fully functional and mature T cells. The corrected cells remained four months after transplantation, indicating that the basic editing had corrected the mutation in the true self-renewing blood stem cells. The results suggest that the corrected blood stem cells could persist over the long term and produce the T cells that patients would need to lead healthy lives. |
The gene editing technique CRISPR/Cas9 has allowed researchers to make precise and impactful changes to an organism's DNA to fix mutations that cause genetic disease.
BigField GEG Tech's insight:
CRISPR/Cas9 method can lead to unintended DNA mutations that can have negative effects. Recently, Japanese researchers have developed a new gene-editing technique that is as effective as CRISPR/Cas9, yet significantly reduces these unintended mutations. In a new study published in Nature Communications , researchers led by Osaka University have introduced a new technique called NICER, based on the creation of several small cuts in single DNA strands by an enzyme Cas 9 nickase. For their first experiments, the research team used human lymphoblastic cells with a known heterozygous mutation in a gene called TK1. When these cells were treated with nickase to induce a single cut in the TK1 region, TK1 activity was recovered at a low rate. However, when nickase induced multiple cuts in this region on both homologous chromosomes, the efficiency of gene correction was increased approximately seventeen-fold via activation of a cellular repair mechanism. Because the NICER method does not involve DNA double-strand breaks or the use of exogenous DNA, this technique appears to be a safe alternative to conventional CRISPR/Cas9 methods.
A broad new strategy could hold hope for treating virtually all blood cancers with CAR T cell therapy, which is currently approved for five subtypes of blood cancer.
BigField GEG Tech's insight:
Until now, researchers have lacked the tools to create a targeted cell therapy approach that could work on all the different forms of blood and bone marrow cancers. A new solution could solve a major problem in immunotherapy, namely the inability to target surface markers present on both cancerous and healthy cells. In the study, published today in Science Translational Medicine, researchers used CAR T cells engineered to target CD45, a surface marker present on almost all blood cells, including almost all blood cancer cells. Since CD45 is also found on healthy blood cells, the research team used CRISPR base editing to develop a method called "epitope editing" to overcome the challenges of an anti-CD45 strategy, which would otherwise result in potentially lethal low blood counts and threatening side effects. The modified version of CD45 still functions, but differs sufficiently from normal CD45 for anti-CD45 CAR T cells not to recognize and attack it. This study lays the foundations for a more universal approach that could potentially extend CAR T cell therapy to all blood cancers.
In the context of relapsed and refractory childhood pre-B cell acute lymphoblastic leukemia (R/R B-ALL), CD19-targeting chimeric antigen receptor (CAR)-T cells often induce durable remissions, which requires the persistence of CAR-T cells. In this study, we systematically analyzed CD19 CAR-T cells of 10 children with R/R B-ALL enrolled in the CARPALL trial via high-throughput single-cell gene expression and T cell receptor sequencing of infusion products and serial blood and bone marrow samples up to 5 years after infusion. We show that long-lived CAR-T cells developed a CD4/CD8 double-negative phenotype with an exhausted-like memory state and distinct transcriptional signature. This persistence signature was dominant among circulating CAR-T cells in all children with a long-lived treatment response for which sequencing data were sufficient (4/4, 100%). The signature was also present across T cell subsets and clonotypes, indicating that persisting CAR-T cells converge transcriptionally. This persistence signature was also detected in two adult patients with chronic lymphocytic leukemia with decade-long remissions who received a different CD19 CAR-T cell product. Examination of single T cell transcriptomes from a wide range of healthy and diseased tissues across children and adults indicated that the persistence signature may be specific to long-lived CAR-T cells. These findings raise the possibility that a universal transcriptional signature of clinically effective, persistent CD19 CAR-T cells exists. In children with relapsed or refractory B cell acute lymphoblastic leukemia and in complete remission after CD19 CAR-T cell therapy, long-lived CAR-T cells express a persistence gene signature that is also present in persistent CD19 CAR-T cells from adults with chronic lymphocytic leukemia.
BigField GEG Tech's insight:
CAR T cells have become an established treatment option for children with a rare form of relapsed or incurable leukemia. One of the key factors determining whether treatment will lead to lasting remission of leukemia is how long the CAR T cells live in the body. One team was able to study the cells of 10 children enrolled in a pioneering clinical trial (CARPALL) for up to five years after their initial CAR T cell treatment. This has enabled them to better understand why some of these CAR T cells remain in a patient's bloodstream, and why others disappear early, potentially allowing the cancer to recur. Using techniques that analyze individual cells at the genetic level to understand what they do, the scientists were able to identify a unique "signature" in long-lived CAR T cells. The signature suggested that long-lived CAR T cells in the blood transformed into a different state that allowed them to continue monitoring the patient's body for cancer cells. As part of the study, the researchers identified key genes in CAR T cells that appeared to enable them to persist in the body for a long time. These genes will provide a starting point for future studies to identify markers of persistence in CAR T-cell products as they are manufactured, and ultimately to improve their efficacy.
Induced pluripotent stem (iPS) cells have a great impact on biology and medicine, and they are expected to improve regenerative medicine.
BigField GEG Tech's insight:
Fabry disease is caused by a genetic deficiency of α-galactosidase A (GLA), leading to the accumulation of its substrates such as globotriaosylceramide and globotriaosylsphingosine. Researchers have therefore developed a modified enzyme, modified α-N-acetylgalactosaminidase (mNAGA), to cure Fabry disease by changing the substrate specificity of NAGA to that of GLA. In this study, researchers tested whether genome-editing transplantation of mNAGA-secreting induced pluripotent stem cells (iPS) cells could deliver GLA activity in vivo. They therefore generated mNAGA-secreting iPS cells by TALEN-mediated knock-in at the AAVS1 site, a refuge locus. Furthermore, to exclude possible immunogenic reactions caused by endogenous GLA from iPS cells in patients, they disrupted the GLA gene by CRISPR-Cas9. When cardiomyocytes and fibroblasts from the Fabry model without GLA activity were co-cultured with mNAGA-secreting iPS cells, GLA activity was restored by mNAGA-expressing cells in vitro. Next, they transplanted the mNAGA-secreting iPS cells into the testes of mouse models of Fabry disease. After 7 or 8 weeks, GLA activity in the liver was significantly improved, although no recovery of activity was observed in the heart, kidneys or blood plasma.
Gene therapy that alters hemoglobin genes may be an answer to curing sickle cell disease (SCD) and beta thalassemia. These two common life-threatening anemias afflict millions of individuals across the globe. Scientists at St. Jude Children's Research Hospital and the Broad Institute of MIT and Harvard used a next-generation genome editing technology, adenosine base editing, to restart fetal hemoglobin expression in SCD patient cells. The approach raised the expression of fetal hemoglobin to higher, more stable, and more uniform levels than other genome editing technologies that use CRISPR/Cas9 nuclease in human hematopoietic stem cells. The findings were published in Nature Genetics.
BigField GEG Tech's insight:
Gene therapy that alters hemoglobin genes may be an answer to curing sickle cell disease (SCD) and beta thalassemia. These two common life-threatening anemias afflict millions of individuals across the globe. Scientists at St. Jude Children's Research Hospital and the Broad Institute of MIT and Harvard used a next-generation genome editing technology, adenosine base editing, to restart fetal hemoglobin expression in SCD patient cells. The approach raised the expression of fetal hemoglobin to higher, more stable, and more uniform levels than other genome editing technologies that use CRISPR/Cas9 nuclease in human hematopoietic stem cells. The findings were published in Nature Genetics.
BigField GEG Tech's insight:
Overall survival for patients with melanoma that has spread to the brain is only four to six months. Researchers have therefore investigated a new therapeutic approach to more effectively target melanoma in the brain. The therapy devised by the scientists uses a "twin stem cell model" designed to maximize an attack on cancer cells that have spread to a part of the brain known as the leptomeninges. A stem cell releases a cancer-killing virus (oncolytic). Using stem cells to deliver the virus amplifies the amount of virus that can be released, and ensures that the virus will not be degraded by circulating antibodies before it is released on cancer cells. However, the oncolytic virus also destroys the cells that release it, making it an unsustainable therapeutic option. Therefore, scientists have used CRISPR/Cas9 gene editing to create a second stem cell that cannot be targeted by the oncolytic virus and instead releases proteins (immunomodulators) that boost the immune system to help fight cancer. Twin stem cells can be delivered by intrathecal injection. Unlike other immunotherapies that have appeared in recent years, it does not need to be administered repeatedly. The researchers point out that this approach can be used in other cancers with metastases.
Vα24-invariant natural killer T cells (NKTs) have anti-tumor properties that can be enhanced by chimeric antigen receptors (CARs). Here we report updated interim results from the first-in-human phase 1 evaluation of autologous NKTs co-expressing a GD2-specific CAR with interleukin 15 (IL15) (GD2-CAR.15) in 12 children with neuroblastoma (NB). The primary objectives were safety and determination of maximum tolerated dose (MTD). The anti-tumor activity of GD2-CAR.15 NKTs was assessed as a secondary objective. Immune response evaluation was an additional objective. No dose-limiting toxicities occurred; one patient experienced grade 2 cytokine release syndrome that was resolved by tocilizumab. The MTD was not reached. The objective response rate was 25% (3/12), including two partial responses and one complete response. The frequency of CD62L+NKTs in products correlated with CAR-NKT expansion in patients and was higher in responders (n = 5; objective response or stable disease with reduction in tumor burden) than non-responders (n = 7). BTG1 (BTG anti-proliferation factor 1) expression was upregulated in peripheral GD2-CAR.15 NKTs and is a key driver of hyporesponsiveness in exhausted NKT and T cells. GD2-CAR.15 NKTs with BTG1 knockdown eliminated metastatic NB in a mouse model. We conclude that GD2-CAR.15 NKTs are safe and can mediate objective responses in patients with NB. Additionally, their anti-tumor activity may be enhanced by targeting BTG1. ClinicalTrials.gov registration: NCT03294954 . In updated results from a phase 1 trial of GD2-specific CAR-NKT cells in patients with neuroblastoma, no dose-limiting toxicities were observed across multiple dose levels; the maximum tolerated dose was not reached; and there was evidence of anti-tumor activity.
BigField GEG Tech's insight:
Researchers report, in Nature Medicine, the interim results of a first-in-man phase 1 clinical trial evaluating the safety, antitumor activity and immunological characteristics of a genetically engineered natural killer (NKT) cell immunotherapy for neuroblastoma, a childhood tumor that most commonly arises in the adrenal gland. NKT cells were engineered to express a GD2-specific CAR, which enables immune cells to target a molecule found on the surface of neuroblastoma cells, and interleukin-15 a natural protein that supports NKT cell survival. Based on results obtained in 12 patients with recurrent stage 4 neuroblastoma resistant to other therapies, the researchers found that the treatment was safe for all 12 patients on four doses. No dose-limiting toxicities were reported. A further discovery revealed a regulatory gene in NKT cells that could have an impact on treatment efficacy. Leveraging the multiomics platform of key collaborator Immunai, Inc. the researchers discovered that up-regulation of the anti-proliferation factor 1 gene BTG in CAR NKT-infused cells indicates cell exhaustion and limits the functional activity of CAR NKT cells. Conversely, artificially reducing BTG1 expression in CAR NKT cells enhanced their therapeutic activity against neuroblastoma in a mouse model
Antibiotic treatments have detrimental effects on the microbiome and lead to antibiotic resistance. To develop a phage therapy against a diverse range of clinically relevant Escherichia coli, we screened a library of 162 wild-type (WT) phages, identifying eight phages with broad coverage of E. coli, complementary binding to bacterial surface receptors, and the capability to stably carry inserted cargo. Selected phages were engineered with tail fibers and CRISPR–Cas machinery to specifically target E. coli. We show that engineered phages target bacteria in biofilms, reduce the emergence of phage-tolerant E. coli and out-compete their ancestral WT phages in coculture experiments. A combination of the four most complementary bacteriophages, called SNIPR001, is well tolerated in both mouse models and minipigs and reduces E. coli load in the mouse gut better than its constituent components separately. SNIPR001 is in clinical development to selectively kill E. coli, which may cause fatal infections in hematological cancer patients. Phage engineered with tail fibers and CRISPR–Cas reduce Escherichia coli load in animals.
BigField GEG Tech's insight:
The emergence of antibiotic-resistant pathogens represents a major global challenge. In numerous disease areas, current medical practice relies on increasingly aggressive therapies, which can sometimes result in patients experiencing life-threatening infections, including those caused by antibiotic-resistant bacteria. SNIPR001's mechanism of action, as a CRISPR-armed phage therapeutic that specifically targets and eradicates E. coli in the gut, is designed to prevent infections from spreading into the bloodstream and represents a promising advancement against antibiotic-resistant pathogens. This publication represents a significant validation of the ground-breaking technology research done at the laboratories and collaborators of SNIPR Biome, the company pioneering CRISPR-based microbial gene therapy, and also opens up the possibility of targeting other pathogens. SNIPR001 has been granted a Fast-Track designation by the US Food and Drug Administration, was supported by CARB-X, and SNIPR is currently conducting a Phase 1 trial in the US to evaluate its safety and efficacy in reducing E. coli in the gut without disturbing the overall gut microbiome.
CRISPR-Cas9 is widely used to edit the genome by studying genes of interest and modifying disease-associated genes.
BigField GEG Tech's insight:
One of the drawbacks of genome editing is that there are growing concerns about mutations and off-target effects. Researchers then hypothesized that current editing protocols that use Cas9 cause excessive DNA cleavage, resulting in some of the mutations. To test this hypothesis, the researchers built a system called "AIMS" in mouse cells, which assessed Cas9 activity separately for each chromosome. Their results showed that the commonly used method was associated with very high editing activity. They determined that this high activity caused some of the undesirable side effects, so they looked for gRNA editing methods that could suppress it. They found that an additional cytosine extension at the 5' end of the gRNA was effective as a "safeguard" against overactivity and controlled DNA cleavage. As a result of this study, the first mathematical model of the correlation between various genome editing patterns and Cas9 activity was created that can maximize the desired editing efficiency by developing activity-regulating gRNAs with appropriate Cas9 activity.
From
www
Decades of oncologic clinical use have demonstrated that cancer immunotherapy provides
BigField GEG Tech's insight:
Immunotherapy is an essential component of cancer treatment. However, despite the success of immunotherapies for various types of cancer, only a few cancer patients have shown responses to current immune therapies and therapies can have adverse effects. Therefore, researchers have created intramuscular messenger ribonucleic acid (mRNA) lipid nanoparticle (LNP) vaccines as likely candidates for therapeutic cancer vaccines. RNA molecules can be encapsulated in LNPs. In addition, single-stranded mRNA can encode tumor vaccine neo-antigens, while small double-stranded interfering RNA can encode knockdown checkpoint inhibitors to adjust immune responses through RNA-induced activation of suppressed immune cells. Circular-type RNA can increase expression time, which benefits the generation of chimeric antigen receptors (CAR) in vivo and vaccine antigens. Targeted delivery of LNP in vivo would be essential for generating CAR-T macrophages and lymphocytes, in vivo. Based on the results of the study, the LNP mRNA vaccine platform could be used to develop next-generation personalized cancer vaccines. However, additional research is needed to further our understanding of cancer biology and improve vaccine design for faster clinical translation.
Chimeric antigen receptor T-cell (CAR-T) therapy has transformed cancer treatment, yet relatively few studies have investigated the impact of the therapy on longitudinal patient quality of life – an aspect of care that often suffers from receiving traditional intensive cancer medications, such as chemotherapy.
BigField GEG Tech's insight:
A new study shows that some effective cancer treatments, such as CAR-T cells, significantly improve quality of life six months after receiving therapy. To conduct the study, researchers recruited 103 patients aged 23 to 90 years with a diagnosis of blood cancer from April 2019 to November 2021. The researchers administered self-reported questionnaires measuring quality of life variables at time intervals including before CAR-T cell infusion and one week, one month, three months, and six months after CAR-T cell infusion. Quality of life was measured using a 27-item questionnaire known as the General Cancer Therapy Functional Assessment, which is composed of four different subscales (physical, functional, emotional, and social). Psychological distress was measured using the Hospital Anxiety and Depression Scale. Finally, major depressive symptoms were measured using the PHQ-9, and symptoms of posttraumatic stress disorder were measured using the Posttraumatic Stress Checklist. While most study participants eventually experienced an improvement in quality of life, approximately 20% of patients experienced persistent physical and psychological symptoms, which at times interfered with their quality of life.
BigField GEG Tech's insight:
Remote and precisely controlled activation of the brain is a fundamental challenge in the development of brain–machine interfaces for neurological treatments. Low-frequency ultrasound stimulation can be used to modulate neuronal activity deep in the brain, especially after expressing ultrasound-sensitive proteins. But so far, no study has described an ultrasound-mediated activation strategy whose spatiotemporal resolution and acoustic intensity are compatible with the mandatory needs of brain–machine interfaces, particularly for visual restoration. Here the authors combined the expression of large-conductance mechanosensitive ion channels with uncustomary high-frequency ultrasonic stimulation to activate retinal or cortical neurons over millisecond durations at a spatiotemporal resolution and acoustic energy deposit compatible with vision restoration. The in vivo sonogenetic activation of the visual cortex generated a behaviour associated with light perception. These findings demonstrate that sonogenetics can deliver millisecond pattern presentations via an approach less invasive than current brain–machine interfaces for visual restoration. |






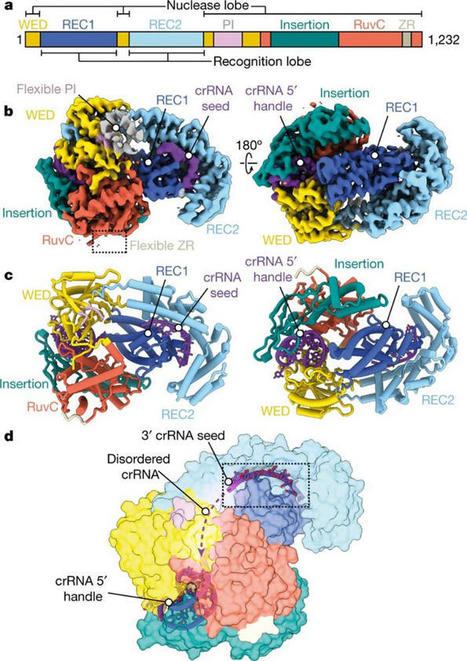









Severe combined immunodeficiency represents rare monogenic disorders with defects in humoral and cellular immunity. RAG genes code for proteins that initiate lymphoid-specific V(D)J recombination, essential for T and B lymphocyte maturation. As a result, individuals carrying pathogenic RAG variants are either completely devoid of, or possess far fewer, B and T cells. In a recent study, researchers described a CRISPR-Cas9-based genome-editing approach to replace the entire respective RAG2 coding sequences (CDS) with a corrective transgene in a 34-positive (CD34+) hematopoietic stem and progenitor cell (HSPC) differentiation cluster. Two recombinant adeno-associated virus serotype 6 (rAAV6) vectors were synthesized to integrate a green fluorescent protein expression cassette and a bovine growth hormone polyA sequence into CD34+ HSPCs after delivery of a single modified RAG2 guide RNA or the Cas9 ribonucleoprotein complex. The researchers' strategy completely replaced RAG2 CDS to treat autosomal recessive immunity disorders. The method induced expression of RAG2 coding DNA without exceeding endogenous RAG2 expression levels. Correction donors promoted the successful recombination and differentiation of V(D)J into CD3 + TCRγδ + and CD3 + TCRαβ + T cells.