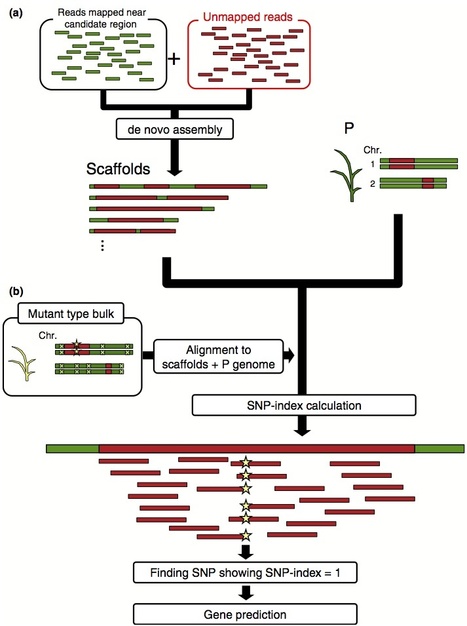
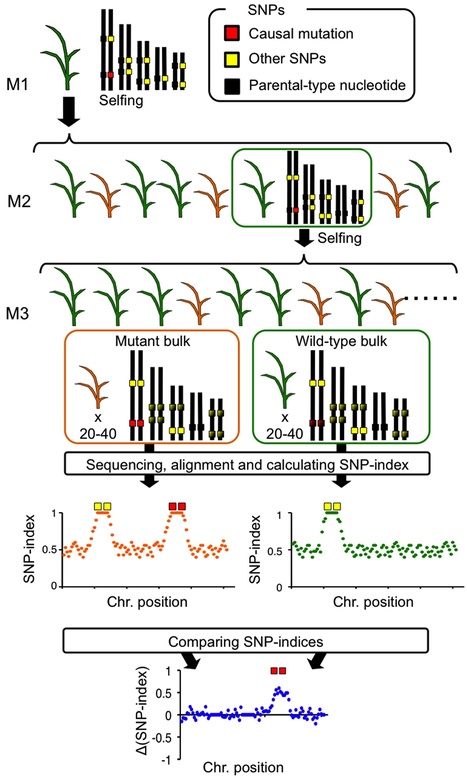
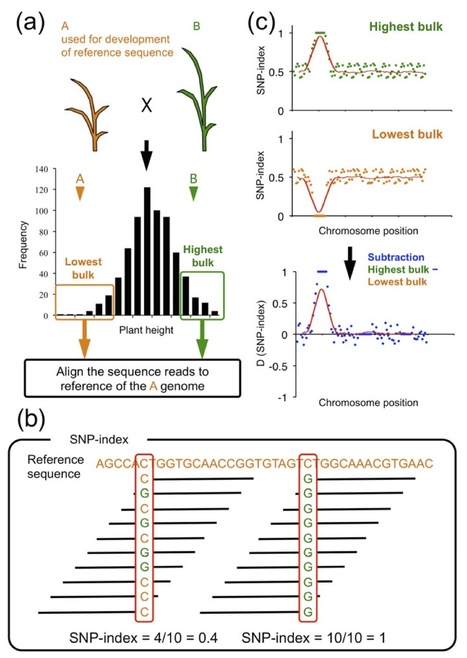
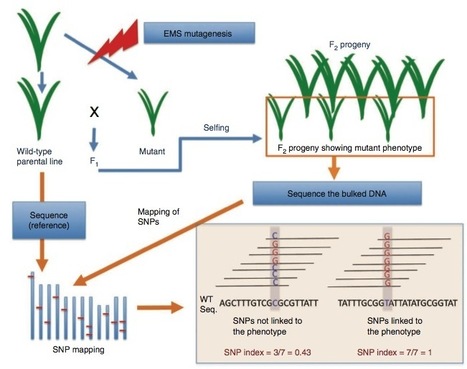
Following the 2011 earthquake and tsunami that affected Japan, >20,000 ha of rice paddy field was inundated with seawater, resulting in salt contamination of the land. As local rice landraces are not tolerant of high salt concentrations, we set out to develop a salt-tolerant rice cultivar. We screened 6,000 ethyl methanesulfonate (EMS) mutant lines of a local elite cultivar, 'Hitomebore', and identified a salt-tolerant mutant that we name hitomebore salt tolerant 1 (hst1). In this Correspondence, we report how we used our MutMap method to rapidly identify a loss-of-function mutation responsible for the salt tolerance of hst1 rice. The salt-tolerant hst1 mutant was used to breed a salt-tolerant variety named 'Kaijin', which differs from Hitomebore by only 201 single-nucleotide polymorphisms (SNPs). Field trials showed that it has the same growth and yield performance as the parental line under normal growth conditions. Notably, production of this salt-tolerant mutant line ready for delivery to farmers took only two years using our approach.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login

Everything about MutMap, a method based on whole-genome sequencing of pooled DNA from a segregating population http://tinyurl.com/mutmap-nbt
Curated by
Kamoun Lab @ TSL
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|