 Your new post is loading...
 Your new post is loading...

|
Scooped by
dromius
|
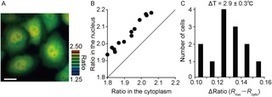
Nakano et al, 2017 Temperature is a fundamental physical parameter that plays an important role in biological reactions and events. Although thermometers developed previously have been used to investigate several important phenomena, such as heterogeneous temperature distribution in a single living cell and heat generation in mitochondria, the development of a thermometer with a sensitivity over a wide temperature range and rapid response is still desired to quantify temperature change in not only homeotherms but also poikilotherms from the cellular level to in vivo. To overcome the weaknesses of the conventional thermometers, such as a limitation of applicable species and a low temporal resolution, owing to the narrow temperature range of sensitivity and the thermometry method, respectively, we developed a genetically encoded ratiometric fluorescent temperature indicator, gTEMP, by using two fluorescent proteins with different temperature sensitivities. Our thermometric method enabled a fast tracking of the temperature change with a time resolution of 50 ms. We used this method to observe the spatiotemporal temperature change between the cytoplasm and nucleus in cells, and quantified thermogenesis from the mitochondria matrix in a single living cell after stimulation with carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, which was an uncoupler of oxidative phosphorylation. Moreover, exploiting the wide temperature range of sensitivity from 5°C to 50°C of gTEMP, we monitored the temperature in a living medaka embryo for 15 hours and showed the feasibility of in vivo thermometry in various living species.

|
Scooped by
dromius
|
Vaa3D is a handy, fast, and versatile 3D/4D/5D Image Visualization & Analysis System for Bioimages & Surface Objects. It also provides many unique functions ... It is also Open Source, supports a very simple and powerful plugin interface and thus can be extended & enhanced easily... Vaa3D is a cross-platform (Mac, Linux, and Windows) tool for visualizing large-scale (gigabytes, and 64-bit data) 3D image stacks and various surface data. It is also a container of powerful modules for 3D image analysis (cell segmentation, neuron tracing, brain registration, annotation, quantitative measurement and statistics, etc) and data management. This makes Vaa3D suitable for various bioimage informatics applications, and a nice platform to develop new 3D image analysis algorithms for high-throughput processing. In short, Vaa3D streamlines the workflow of visualization-assisted analysis. In our latest development, Vaa3D can render 5D (spatial-temporal) data directly in 3D volume-rendering mode; it supports convenient and interactive local and global 3D views at different scales... it even has a Matlab file IO toolbox. Probably most importantly, you can now write your own plugins to take advantage of the Vaa3D platform!

|
Scooped by
dromius
|
(via T. Lahaye, thanks) Osborn et al, 2013 Recessive dystrophic epidermolysis bullosa (RDEB) is characterized by a functional deficit of type VII collagen protein due to gene defects in the type VII collagen gene (COL7A1). Gene augmentation therapies are promising, but run the risk of insertional mutagenesis. To abrogate this risk, we explored the possibility of using engineered transcription activator-like effector nucleases (TALEN) for precise genome editing. We report the ability of TALEN to induce site-specific double-stranded DNA breaks (DSBs) leading to homology-directed repair (HDR) from an exogenous donor template. This process resulted in COL7A1 gene mutation correction in primary fibroblasts that were subsequently reprogrammed into inducible pluripotent stem cells and showed normal protein expression and deposition in a teratoma-based skin model in vivo. Deep sequencing-based genome-wide screening established a safety profile showing on-target activity and three off-target (OT) loci that, importantly, were at least 10 kb from a coding sequence. This study provides proof-of-concept for TALEN-mediated in situ correction of an endogenous patient-specific gene mutation and used an unbiased screen for comprehensive TALEN target mapping that will cooperatively facilitate translational application.

|
Scooped by
dromius
|
Fluorescent fusion protein knockout mediated by anti-GFP nanobody - Nature Struct. Mol. Biology
http://www.nature.com/nsmb/journal/v19/n1/full/nsmb.2180.html Caussinus et al (2012) The use of genetic mutations to study protein functions in vivo is a central paradigm of modern biology. Recent advances in reverse genetics such as RNA interference and morpholinos are widely used to further apply this paradigm. Nevertheless, such systems act upstream of the proteic level, and protein depletion depends on the turnover rate of the existing target proteins. Here we present deGradFP, a genetically encoded method for direct and fast depletion of target green fluorescent protein (GFP) fusions in any eukaryotic genetic system. This method is universal because it relies on an evolutionarily highly conserved eukaryotic function, the ubiquitin pathway. It is traceable, because the GFP tag can be used to monitor the protein knockout. In many cases, it is a ready-to-use solution, as GFP protein-trap stock collections are being generated in Drosophila melanogaster and in Danio rerio.

|
Scooped by
dromius
|
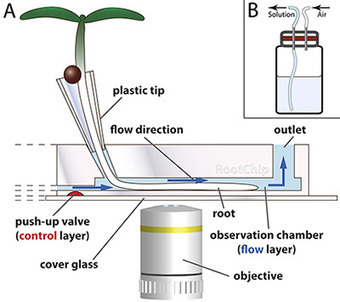
This article provides a protocol for cultivation of Arabidopsis seedlings in the RootChip, a microfluidic imaging platform that combines automated control of growth conditions with microscopic root monitoring and FRET-based measurement

|
Scooped by
dromius
|
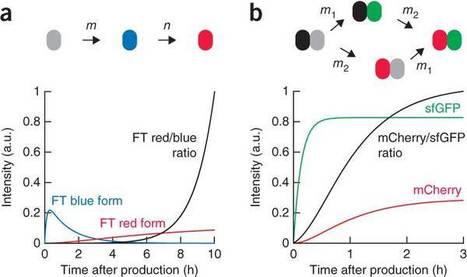
The functional state of a cell is largely determined by the spatiotemporal organization of its proteome. Technologies exist for measuring particular aspects of protein turnover and localization, but comprehensive analysis of protein dynamics across different scales is possible only by combining several methods. Here we describe tandem fluorescent protein timers (tFTs), fusions of two single-color fluorescent proteins that mature with different kinetics, which we use to analyze protein turnover and mobility in living cells. We fuse tFTs to proteins in yeast to study the longevity, segregation and inheritance of cellular components and the mobility of proteins between subcellular compartments; to measure protein degradation kinetics without the need for time-course measurements; and to conduct high-throughput screens for regulators of protein turnover. Our experiments reveal the stable nature and asymmetric inheritance of nuclear pore complexes and identify regulators of N-end rule-mediated protein degradation.

|
Scooped by
dromius
|
Our objective was to test whether or not plastids and mitochondria, the two DNA-containing organelles, move between cells in plants. As our experimental approach, we grafted two different species of tobacco, Nicotiana tabacum and Nicotiana sylvestris. Grafting triggers formation of new cell-to-cell contacts, creating an opportunity to detect cell-to-cell organelle movement between the genetically distinct plants. We initiated tissue culture from sliced graft junctions and selected for clonal lines in which gentamycin resistance encoded in the N. tabacum nucleus was combined with spectinomycin resistance encoded in N. sylvestris plastids. Here, we present evidence for cell-to-cell movement of the entire 161-kb plastid genome in these plants, most likely in intact plastids. We also found that the related mitochondria were absent, suggesting independent movement of the two DNA-containing organelles. Acquisition of plastids from neighboring cells provides a mechanism by which cells may be repopulated with functioning organelles. Our finding supports the universality of intercellular organelle trafficking and may enable development of future biotechnological applications.

|
Scooped by
dromius
|
We used bimolecular fluorescence complementation technology to screen a plant cDNA library against a bait protein directly in plants. As proof of concept, we used the N-terminal fragment of yellow fluorescent protein– or nVenus-tagged Agrobacterium tumefaciens VirE2 and VirD2 proteins and the C-terminal extension (CTE) domain of Arabidopsis thaliana telomerase reverse transcriptase as baits to screen an Arabidopsis cDNA library encoding proteins tagged with the C-terminal fragment of yellow fluorescent protein. A library of colonies representing ∼2 × 105 cDNAs was arrayed in 384-well plates. Sequential screening of subsets of cDNAs in Arabidopsis leaf or tobacco (Nicotiana tabacum) Bright Yellow-2 protoplasts identified single cDNA clones encoding proteins that interact with either, or both, of the Agrobacterium bait proteins, or with CTE. T-DNA insertions in the genes represented by some cDNAs revealed five novel Arabidopsis proteins important for Agrobacterium-mediated plant transformation. We also used this cDNA library to confirm VirE2-interacting proteins in orchid (Phalaenopsis amabilis) flowers. Thus, this technology can be applied to several plant species.

|
Scooped by
dromius
|
Transient gene expression, in plant protoplasts or specific plant tissues, is a key technique in plant molecular cell biology, aimed at exploring gene products and their modifications to examine functional subdomains, their interactions with other biomolecules, and their subcellular localization. Here, we highlight some of the major advantages and potential pitfalls of the most commonly used transient gene expression models and illustrate how ectopic expression and the use of dominant mutants can provide insights into protein function.

|
Scooped by
dromius
|
Automated cell segmentation in time-lapse confocal images of growing Arabidopsis combined with ratiometric imaging of fluorescent gene expression reporters permits the correlation of cellular properties with gene expression in the growing plant.

|
Scooped by
dromius
|
Our objective was to functionalize diatom biosilica with a reagent-less sensor dependent on ligand-binding and conformational change to drive FRET-based signaling capabilities. A fusion protein designed to confer such properties included a bacterial periplasmic ribose binding protein (R) flanked by CyPet (C) and YPet (Y), cyan and yellow fluorescent proteins that act as a FRET pair.This work demonstrated that the nano-architecture of the genetically modified biosilica cell wall was able to support the functionality of the relatively complex Sil3-CyPet-RBP-YPet fusion protein with its requirement for ligand-binding and conformational change for FRET-signal generation.

|
Scooped by
dromius
|
Nature Methods - Spatiotemporal control of gene expression by a light-switchable transgene system

|
Scooped by
dromius
|
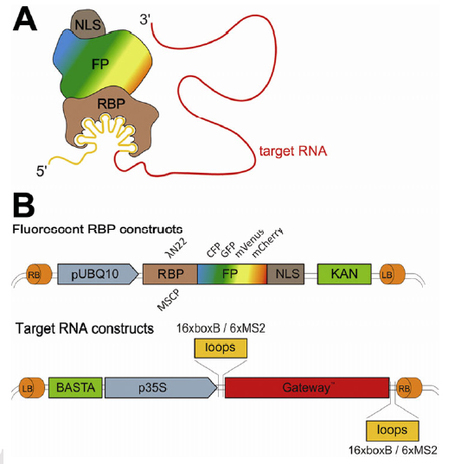
Schoenberger et al, 2012, Plant Journal Localisation of RNA, a mechanism for controlling translation in a spatial and temporal fashion requires processing and assembly of RNA into transport granules in the nucleus, transport towards cytoplasmic destinations and regulation of its activity. Compared with animal model systems little is known about RNA dynamics and motility in plants. Here, we introduce the λN22 RNA stem-loop binding system for the in vivo visualization of RNA in plant cells. The λN22 system consists of two-components, the λN22 RNA binding peptide and the corresponding boxB stem-loops. We generated fusions of λN22 to different fluorophores and a GATEWAY vector series for simple fusion of any target RNA 5’ or 3’ to boxB stem-loops. We show that the λN22 system can be used to detect RNAs in transient expression assays and that it offers advantages compared to the previously described MS2 system.
|

|
Scooped by
dromius
|
Protein networks and signaling cascades are key mechanisms for intra- and intercellular signal transduction. Identifying the interacting partners of a protein can provide vital clues regarding its physiological role. The Bimolecular Fluorescence Complementation (BiFC) assay has become a routine tool for in vivo analysis of protein-protein interactions and their subcellular location. Although the BiFC system has improved since its inception, the available options for in planta analysis are still subject to very low signal to noise ratios, and a systematic comparison of BiFC confounding background signals has been lacking. Background signals can obscure weak interactions, provide false positives, and decrease confidence in true positives. To overcome these problems, we performed an extensive in planta analysis of published BiFC fragments used in metazoa and plants, and then developed an optimized single vector BiFC system which utilizes monomeric Venus (mVenus) split at residue 210, and contains an integrated mTurquoise2 marker to precisely identify transformed cells in order to distinguish true negatives. Here we provide our streamlined Double ORF Expression (pDOE) BiFC system, and show that our advance in BiFC methodology functions even with an internally fused mVenus210 fragment. We illustrate the efficacy of the system by providing direct visualization of Arabidopsis MLO1 interacting with a calmodulin-like (CML) protein, and by showing that heterotrimeric G-protein subunits Gα (GPA1) and Gβ (AGB1) interact in plant cells. We further demonstrate that GPA1 and AGB1 each physically interact with PLDα1 in planta, and that mutation of the so-called PLDα1 “DRY” motif abolishes both of these interactions.

|
Scooped by
dromius
|
Website presenting the available plant image analysis softwares

|
Scooped by
dromius
|
A 993-bp regulatory region upstream of the translation start codon of subtilisin-like serine protease gene was isolated from Gossypium barbadense. This (T/A)AAAG-rich region, GbSLSP, and its 5′- and 3′-truncated versions were transferred into tobacco and Arabidopsis after fusing with GUS or GFP. Histochemical and quantitative GUS analysis and confocal GFP fluorescence scanning in the transgenic plants showed that the GbSLSP-driven GUS and GFP expressed preferentially in guard cells, whereas driven by GbSLSPF2 to GbSLSPF4, the 5′-truncated GbSLSP versions with progressively reduced Dof1 elements, both GUS and GFP expressed exclusively in guard cells, and the expression strength declined with (T/A)AAAG copy decrement. Deletion of 5′-untranslated region from GbSLSP markedly weakened the activity of GUS and GFP, while deletion from the strongest guard cell-specific promoter, GbSLSPF2, not only significantly decreased the expression strength, but also completely abolished the guard cell specificity. These results suggested both guard cell specificity and expression strength of the promoters be coordinately controlled by 5′-untranslated region and a cluster of at least 3 (T/A)AAAG elements within a region of about 100 bp relative to transcription start site. Our guard cell-specific promoters will enrich tools to manipulate gene expression in guard cells for scientific research and crop improvement.

|
Scooped by
dromius
|
Check out the apoptosis movie - cool stuff!

|
Scooped by
dromius
|
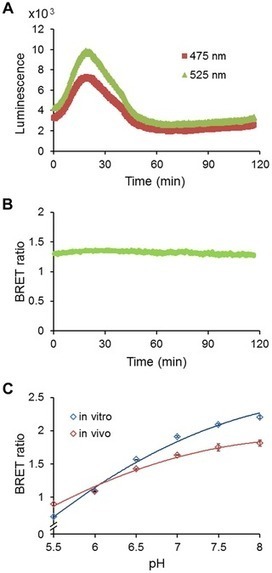
We report the development of a genetically encodable and ratiometic pH probe named “pHlash” that utilizes Bioluminescence Resonance Energy Transfer (BRET) rather than fluorescence excitation. The pHlash sensor–composed of a donor luciferase that is genetically fused to a Venus fluorophore–exhibits pH dependence of its spectral emission in vitro. When expressed in either yeast or mammalian cells, pHlash reports basal pH and cytosolic acidification in vivo. Its spectral ratio response is H+ specific; neither Ca++, Mg++, Na+, nor K+ changes the spectral form of its luminescence emission. Moreover, it can be used to image pH in single cells. This is the first BRET-based sensor of H+ ions, and it should allow the approximation of pH in cytosolic and organellar compartments in applications where current pH probes are inadequate.

|
Scooped by
dromius
|
An article, which reminds us that imaging itself can influence our beloved plant cells! "In most terrestrial plants, stomata open during the day to maximize the update of CO2 for photosynthesis, but they close at night to minimize water loss. Blue light, among several environmental factors, controls this process. Stomata response to diverse stimuli seems to be dictated by the behaviour of neighbour stomata creating leaf areas of coordinated response. Here individual stomata of Arabidopsis leaves were illuminated with a short blue-light pulse by focusing a confocal argon laser. Beautifully, the illuminated stomata open their pores, whereas their dark-adapted neighbours unexpectedly experience no change. This induction of individual stomata opening by low fluence rates of blue light was disrupted in the phototropin1 phototropin2 (phot1 phot2) double mutant, which exhibits insensitivity of stomatal movements in blue-illuminated epidermal strips. The irradiation of all epidermal cells making direct contact with a given stoma in both wild type and phot1 phot2 plants does not trigger its movement. These results unravel the stoma autonomous function in the blue light response and illuminate the implication of PHOT1 and/or PHOT2 in such response. The micro spatial heterogeneity that solar blue light suffers in partially shaded leaves under natural conditions highlights the physiological significance of the autonomous stomatal behaviour."
"Icy: The open source community software for bio-imaging Over the past decades the image analysis community has put substantial efforts into developing algorithms for various applications, yet their visibility often remained clustered to a handful of people, due to lack of proper means of communication and advertising. Meanwhile, numerous experts in the biology community have been turning to image analysis to answer their questions, and yet often fail to find adapted (or affordable) tools to fit their needs.
Icy provides an integrated platform that aims at bridging the gap between developers and users, by combining: a) an open-source image analysis software, offering a powerful and flexible environment for developers such as applied mathematicians to write algorithms fast and efficiently; b) a common set of tools to view and manipulate data, and a set of plugins to perform specific quantification or analysis on images; c) a community-based website centralizing all plugins and resources to facilitate their management and maximize their visibility towards users. "
Via Schattat Lab

|
Scooped by
dromius
|
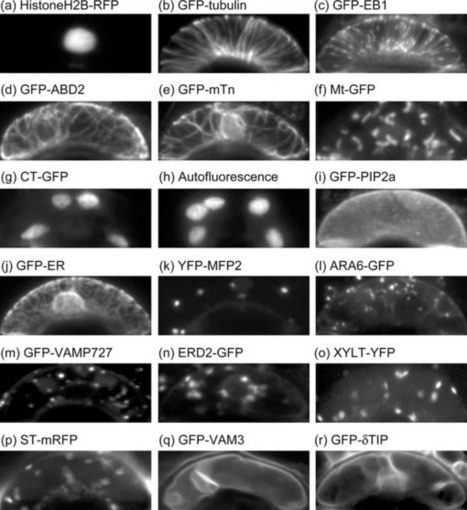
To comprehensively grasp cell biological events in plant stomatal movement, we have captured microscopic images of guard cells with various organelles markers. The 28,530 serial optical sections of 930 pairs of Arabidopsis guard cells have been released as a new image database, named Live Images of Plant Stomata (LIPS). http://hasezawa.ib.k.u-tokyo.ac.jp/lips/images.html

|
Scooped by
dromius
|
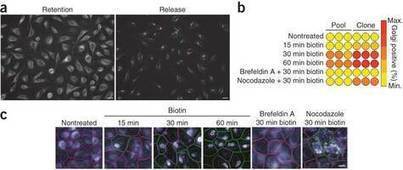
The biotin-reversible interaction between a 'hook' protein localized to a particular cellular compartment and a reporter protein of interest is exploited in a simple system to synchronize protein traffic through the secretory pathway.

|
Scooped by
dromius
|
Stroma-filled tubules named stromules are sporadic extensions of plastids. Earlier, photobleaching was used to demonstrate fluorescent protein diffusion between already interconnected plastids and formed the basis for suggesting that all plastids are able to form networks for exchanging macromolecules. However, a critical appraisal of literature shows that this conjecture is not supported by unequivocal experimental evidence. Here, using photoconvertible mEosFP, we created color differences between similar organelles that enabled us to distinguish clearly between organelle fusion and nonfusion events. Individual plastids, despite conveying a strong impression of interactivity and fusion, maintained well-defined boundaries and did not exchange fluorescent proteins. Our observations provide succinct evidence to refute the long-standing dogma of interplastid connectivity.

|
Scooped by
dromius
|
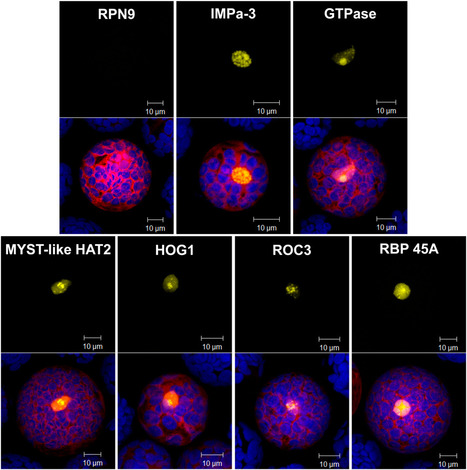
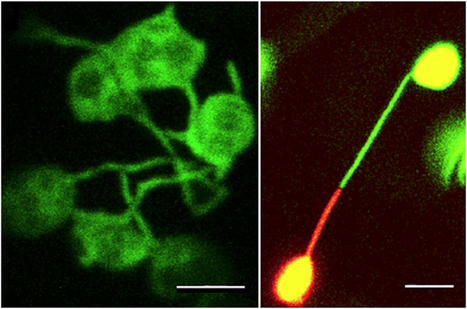
Polyubiquitin chain topology is thought to direct modified substrates to specific fates, but this function-topology relationship is poorly understood, as are the dynamics and subcellular locations of specific polyubiquitin signals. Here we present a general strategy to track linkage-specific polyubiquitin signals ... and to probe their functions. We designed several high-affinity Lys63 polyubiquitin-binding proteins and demonstrate their specificity in vitro and in cells. We apply these tools as competitive inhibitors to dissect the polyubiquitin-linkage dependence of NF-κB activation in several cell types, inferring the essential role of Lys63 polyubiquitin for signaling via the IL-1β and TNF-related weak inducer of apoptosis but not TNF-α receptors. We anticipate live-cell imaging, proteomic and biochemical applications for these tools and extension of the design strategy to other polymeric ubiquitin-like protein modifications.

|
Scooped by
dromius
|
Nature - A novel sensor to map auxin response and distribution at high spatio-temporal resolution
http://go.nature.com/CvhDNj Auxin is a key plant morphogenetic signal but tools to analyse dynamically its distribution and signalling during development are still limited. Auxin perception directly triggers the degradation of Aux/IAA repressor proteins. Here we describe a novel Aux/IAA-based auxin signalling sensor termed DII-VENUS that was engineered in the model plant Arabidopsis thaliana. The VENUS fast maturing form of yellow fluorescent protein7 was fused in-frame to the Aux/IAA auxin-interaction domain (termed domain II; DII) and expressed under a constitutive promoter. We initially show that DII-VENUS abundance is dependent on auxin, its TIR1/AFBs co-receptors and proteasome activities. Next, we demonstrate that DII-VENUS provides a map of relative auxin distribution at cellular resolution in different tissues. DII-VENUS is also rapidly degraded in response to auxin and we used it to visualize dynamic changes in cellular auxin distribution successfully during two developmental responses, the root gravitropic response and lateral organ production at the shoot apex. Our results illustrate the value of developing response input sensors such as DII-VENUS to provide high-resolution spatio-temporal information about hormone distribution and response during plant growth and development.
|


 Your new post is loading...
Your new post is loading...








![Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling [Nat Methods. 2012] | Plant Cell Biology and Microscopy | Scoop.it](https://img.scoop.it/KKHnxuL4BndQqAkIhViVEzl72eJkfbmt4t8yenImKBVvK0kTmF0xjctABnaLJIm9)





