 Your new post is loading...
 Your new post is loading...
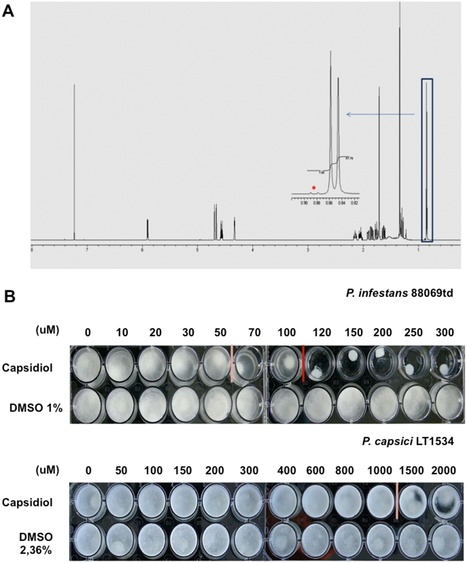
Plants protect themselves against a variety of invading pathogenic organisms via sophisticated defence mechanisms. These responses include deployment of specialized antimicrobial compounds, such as phytoalexins, that rapidly accumulate at pathogen infection sites. However, the extent to which these compounds contribute to species-level resistance and their spectrum of action remain poorly understood. Capsidiol, a defense related phytoalexin, is produced by several solanaceous plants including pepper and tobacco during microbial attack. Interestingly, capsidiol differentially affects growth and germination of the oomycete pathogensPhytophthora infestans and Phytophthora capsici, although the underlying molecular mechanisms remain unknown. In this study we revisited the differential effect of capsidiol on P. infestans and P. capsici, using highly pure capsidiol preparations obtained from yeast engineered to express the capsidiol biosynthetic pathway. Taking advantage of transgenicPhytophthora strains expressing fluorescent markers, we developed a fluorescence-based method to determine the differential effect of capsidiol on Phytophtora growth. Using these assays, we confirm major differences in capsidiol sensitivity between P. infestans and P. capsiciand demonstrate that capsidiol alters the growth behaviour of both Phytophthora species. Finally, we report intraspecific variation within P. infestans isolates towards capsidiol tolerance pointing to an arms race between the plant and the pathogens in deployment of defence related phytoalexins.
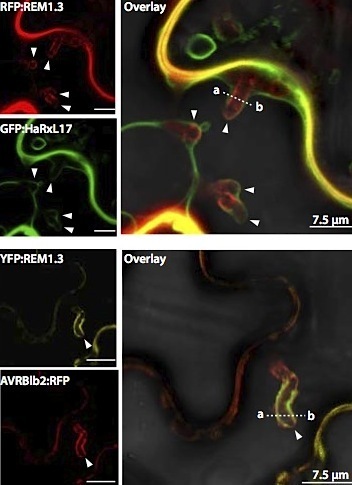
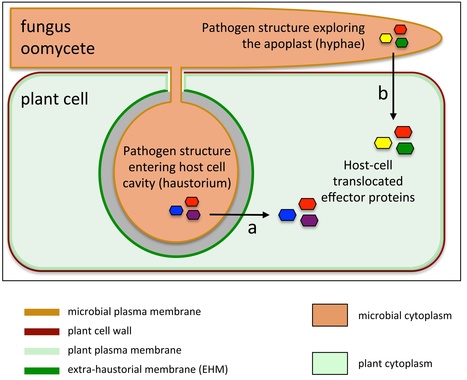
Filamentous plant pathogens such as the late blight pathogenPhytophthora infestans form digit-like infection structures called haustoria inside plant cells. Haustoria enable the pathogen to feed on its host, and secrete effector proteins that modulate the physiology of the host cell to facilitate infection. Haustoria are enveloped by a specialized plant-derived membrane (the extrahaustorial membrane) the biogenesis of which is poorly understood. In this issue, Bozkurt et al. http://www.plantphysiol.org/content/165/3/1005 used the plant membrane microdomain protein REMORIN1.3, known to accumulate around P. infestans haustoria, to reveal discrete extrahaustorial domains labeled by REMORIN1.3 and P. infestans effector AVRblb2. SYNAPTOTAGMIN1, another previously identified perihaustorial protein, localized to subdomains which are mainly not labeled by REMORIN1.3 and AVRblb2. Functional characterization of REMORIN1.3 revealed that it is a susceptibility factor that promotes infection by P. infestans. This activity, and REMORIN1.3 recruitment to the EHM, require REM1.3 membrane-binding domain. These results implicate REMORIN1.3 membrane microdomains in plant susceptibility to an oomycete pathogen. The cover shows Nicotiana benthamiana epidermal cells expressing fluorescently labeled REMORIN1.3 (blue) infected by P. infestans expressing the red fluorescent protein (red). Cover image credits: Sylvain Raffaele.
- Nonhost resistance (NHR) is a plant immune response to resist most pathogens. The molecular basis of NHR is poorly understood, but recognition of pathogen effectors by immune receptors, a response known as effector-triggered immunity, has been proposed as a component of NHR.
- We performed transient expression of 54 Phytophthora infestansRXLR effectors in pepper (Capsicum annuum) accessions. We used optimized heterologous expression methods and analyzed the inheritance of effector-induced cell death in an F2 population derived from a cross between two pepper accessions.
- Pepper showed a localized cell death response upon inoculation with P. infestans, suggesting that recognition of effectors may contribute to NHR in this system. Pepper accessions recognized as many as 36 effectors. Among the effectors, PexRD8 and Avrblb2 induced cell death in a broad range of pepper accessions. Segregation of effector-induced cell death in an F2 population derived from a cross between two pepper accessions fit 15 : 1, 9 : 7 or 3 : 1 ratios, depending on the effector.
- Our genetic data suggest that a single or two independent/complementary dominant genes are involved in the recognition of RXLR effectors. Multiple loci recognizing a series of effectors may underpin NHR of pepper to P. infestans and confer resistance durability.
Filamentous pathogens such as the oomycete Phytophthora infestans infect plants by developing specialized structures termed haustoria inside the host cells. Haustoria are thought to enable secretion of effector proteins into the plant cells. Haustorium biogenesis is therefore critical for pathogen accommodation in the host tissue. Haustoria are enveloped by a specialized host-derived membrane, the extrahaustorial membrane (EHM), which is distinct from the plant plasma membrane. The mechanisms underlying the biogenesis of the EHM are unknown. Remarkably, several plasma membrane localised proteins are excluded from the EHM but the remorin REM1.3 accumulates around P. infestans haustoria. Here, we used overexpression, co-localization with reporter proteins, and super-resolution microscopy in cells infected by P. infestans to reveal discrete EHM domains labelled by REM1.3 and P. infestans effector AVRblb2. Moreover, SYT1 synaptotagmin, another previously identified perihaustorial protein, localized to subdomains which are mainly not labelled by REM1.3 and AVRblb2. Functional characterization of REM1.3 revealed that it is a susceptibility factor that promotes infection by P. infestans. This activity, and REM1.3 recruitment to the EHM, require REM1.3 membrane binding domain. Our results implicate REM1.3 membrane micro-domains in plant susceptibility to an oomycete pathogen.
Since the dawn of agriculture, plant pathogens and pests have been a scourge of humanity. Yet we have come a long way since the Romans attempted to mitigate the effects of plant disease by worshipping and honoring the god Robigus. Books in the Middle Ages by Islamic and European scholars described various plant diseases and even proposed particular disease management strategies. Surprisingly, the causes of plant diseases remained a matter of debate over a long period. It took Henri-Louis Duhamel du Monceau's elegant demonstration in his 1728 “Explication Physique” paper that a “contagious” fungus was responsible for a saffron crocus disease to usher in an era of documented scientific inquiry. Confusion and debate about the exact nature of the causal agents of plant diseases continued until the 19th century, which not only saw the first detailed analyses of plant pathogens but also provided much-needed insight into the mechanisms of plant disease. An example of this is Anton de Bary's demonstration that a “fungus” is a cause, not a consequence, of plant disease. This coming of age of plant pathology was timely. In the 19th century, severe plant disease epidemics hit Europe and caused economic and social upheaval. These epidemics were not only widely covered in the press but also recognized as serious political issues by governments. Many of the diseases, including late blight of potato, powdery and downy mildew of grapevine, as well as phylloxera, were due to exotic introductions from the Americas and elsewhere. These and subsequent epidemics motivated scientific investigations into crop breeding and plant disease management that developed into modern plant pathology science over the 20th century.
Nowadays, our understanding of plant pathogens and the diseases they cause greatly benefits from molecular genetics and genomics. All aspects of plant pathology, from population biology and epidemiology to mechanistic research, are impacted. The polymerase chain reaction (PCR) first enabled access to plant pathogen DNA sequences from historical specimens deposited in herbaria. Historical records in combination with herbarium specimens have turned out to provide powerful tools for understanding the course of past plant epidemics. Recently, thanks to new developments in DNA sequencing technology, it has become possible to reconstruct the genomes of plant pathogens in herbaria. In this article, we first summarize how whole genome analysis of ancient DNA has been recently used to reconstruct the 19th-century potato-blight epidemic that rapidly spread throughout Europe and triggered the Irish potato famine. We then discuss the exciting prospects offered by the emergence of the discipline of ancient plant pathogen genomics.
Both plants and animals rely on nucleotide-binding domain and leucine-rich repeat-containing proteins (NB-LRRs or NLRs) to respond to invading pathogens and activate immune responses. How plant NB-LRR proteins respond to pathogens is poorly understood. We undertook a gain-of-function random mutagenesis screen of the potato NB-LRR immune receptor R3a to study how this protein responds to the effector protein AVR3a from the oomycete pathogen Phytophthora infestans. R3a response can be extended to the stealthy AVR3aEM isoform of the effector while retaining recognition of AVR3aKI. Each one of 8 single amino acid mutations is sufficient to expand the R3a response to AVR3aEM and other AVR3a variants. These mutations occur across the R3a protein, from the N-terminus to different regions of the LRR domain. Further characterization of these R3a mutants revealed that at least one of them was sensitized, exhibiting a stronger response than the wild-type R3a protein to AVR3aKI. Remarkably, the N336Y mutation, near the R3a nucleotide-binding pocket, conferred response to the effector protein PcAVR3a4 from the vegetable pathogen Phytophthora capsici. This work contributes to understanding how NB-LRR receptor specificity can be modulated. Together with knowledge of pathogen effector diversity, this strategy can be exploited to develop synthetic immune receptors.
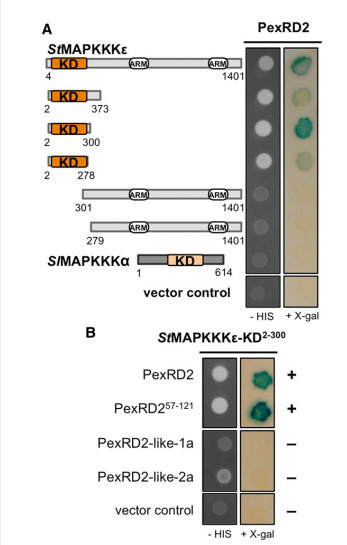
Mitogen-activated protein kinase cascades are key players in plant immune signaling pathways, transducing the perception of invading pathogens into effective defense responses. Plant pathogenic oomycetes, such as the Irish potato famine pathogen Phytophthora infestans, deliver RXLR effector proteins to plant cells to modulate host immune signaling and promote colonization. Our understanding of the molecular mechanisms by which these effectors act in plant cells is limited. Here, we report that the P. infestans RXLR effector PexRD2 interacts with the kinase domain of MAPKKKε, a positive regulator of cell death associated with plant immunity. Expression of PexRD2 or silencing MAPKKKε inNicotiana benthamiana enhances susceptibility to P. infestans. We show that PexRD2 perturbs signaling pathways triggered by or dependent on MAPKKKε. By contrast, homologs of PexRD2 from P. infestans had reduced or no interaction with MAPKKKε and did not promote disease susceptibility. Structure-led mutagenesis identified PexRD2 variants that do not interact with MAPKKKε and fail to support enhanced pathogen growth or perturb MAPKKKε signaling pathways. Our findings provide evidence that P. infestans RXLR effector PexRD2 has evolved to interact with a specific host MAPKKK to perturb plant immunity–related signaling.
Via Suayib Üstün, Kamoun Lab @ TSL
Accelerated gene evolution is a hallmark of pathogen adaptation following a host jump. Here, we describe the biochemical basis of adaptation and specialization of a plant pathogen effector after its colonization of a new host. Orthologous protease inhibitor effectors from the Irish potato famine pathogen, Phytophthora infestans, and its sister species, Phytophthora mirabilis, which is responsible for infection of Mirabilis jalapa, are adapted to protease targets unique to their respective host plants. Amino acid polymorphisms in both the inhibitors and their target proteases underpin this biochemical specialization. Our results link effector specialization to diversification and speciation of this plant pathogen.
Most fruits in our daily diet are the products of domestication and breeding. Here we report a map of genome variation for a major fruit that encompasses ~3.6 million variants, generated by deep resequencing of 115 cucumber lines sampled from 3,342 accessions worldwide. Comparative analysis suggests that fruit crops underwent narrower bottlenecks during domestication than grain crops. We identified 112 putative domestication sweeps; 1 of these regions contains a gene involved in the loss of bitterness in fruits, an essential domestication trait of cucumber. We also investigated the genomic basis of divergence among the cultivated populations and discovered a natural genetic variant in a β-carotene hydroxylase gene that could be used to breed cucumbers with enhanced nutritional value. The genomic history of cucumber evolution uncovered here provides the basis for future genomics-enabled breeding.
Targeted genome engineering (also known as genome editing) has emerged as an alternative to classical plant breeding and transgenic (GMO) methods to improve crop plants. Until recently, available tools for introducing site-specific double strand DNA breaks were restricted to zinc finger nucleases (ZFNs) and TAL effector nucleases (TALENs). However, these technologies have not been widely adopted by the plant research community due to complicated design and laborious assembly of specific DNA binding proteins for each target gene. Recently, an easier method has emerged based on the bacterial type II CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated) immune system. The CRISPR/Cas system allows targeted cleavage of genomic DNA guided by a customizable small noncoding RNA, resulting in gene modifications by both non-homologous end joining (NHEJ) and homology-directed repair (HDR) mechanisms. In this review we summarize and discuss recent applications of the CRISPR/Cas technology in plants.
Sustainable intensification of crop production is essential to ensure food demand is matched by supply as the human population continues to increase. This will require high-yielding crop varieties that can be grown sustainably with fewer inputs on less land. Both plant breeding and genetic modification (GM) methods make valuable contributions to varietal improvement, but targeted genome engineering promises to be critical to elevating future yields. Most such methods require targeting DNA breaks to defined locations followed by either nonhomologous end joining (NHEJ) or homologous recombination. Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can be engineered to create such breaks, but these systems require two different DNA binding proteins flanking a sequence of interest, each with a C-terminal FokI nuclease module. We report here that the bacterial clustered, regularly interspaced, short palindromic repeats (CRISPR) system, comprising a CRISPR-associated (Cas)9 protein and an engineered single guide RNA (sgRNA) that specifies a targeted nucleic acid sequence, is applicable to plants to induce mutations at defined loci.
Recent pathogenomic research on plant parasitic oomycete effector function and plant host responses has resulted in major conceptual advances in plant pathology, which has been possible thanks to the availability of genome sequences.
Few microorganisms match the impact that the oomycetes have had on mankind. This distinct lineage of eukaryotes is well-known for its most notorious member, Phytophthora infestans, the agent of the nineteenth century Irish potato famine, and several other devastating pathogens of cultivated and wild plants [1]. Indeed, more than 60% of oomycete species infect plants [2]. Less known is the fact that many oomycetes are parasitic on animals, from freshwater fish and crustaceans to mammals, such as livestock, pets, and humans [3]. Animal parasitic oomycetes have received much less attention than their plant pathogenic kin, and our understanding of their virulence mechanisms is rudimentary. However, research momentum is poised to accelerate with the first report of the genome of an animal parasitic oomycete. In this issue of PLOS Genetics, Jiang et al. [4] describe the 63 Mbp genome sequence of the fish pathogen Saprolegnia parasitica and highlight a distinct repertoire of candidate virulence genes.
|
Oomycetes form a deep lineage of eukaryotic organisms that includes a large number of plant pathogens that threaten natural and managed ecosystems. We undertook a survey to query the community for their ranking of plant pathogenic oomycete species based on scientific and economic importance. In total, we received 263 votes from 62 scientists in 15 countries for a total of 33 species. The Top 10 species and their ranking are: (1) Phytophthora infestans; (2, tied) Hyaloperonospora arabidopsidis; (2, tied) Phytophthora ramorum; (4) Phytophthora sojae; (5) Phytophthora capsici; (6) Plasmopara viticola; (7) Phytophthora cinnamomi; (8, tied) Phytophthora parasitica; (8, tied) Pythium ultimum; and (10) Albugo candida. The article provides an introduction to these 10 taxa and a snapshot of current research. We hope that the list will serve as a benchmark for future trends in oomycete research.
See also [link below]:
Top 10 plant-parasitic nematodes in molecular plant pathology
Top 10 plant viruses in molecular plant pathology
Top 10 plant pathogenic bacteria in molecular plant pathology
The Top 10 fungal pathogens in molecular plant pathology
http://onlinelibrary.wiley.com/journal/10.1111/(ISSN)1364-3703/homepage/free_poster.htm
The world's human population continues to expand and is predicted to reach ~9 billion by 2040, up from its current level of just over 7 billion. Some estimate that with this rate of population growth, accommodating the increased demand for food will require the world's agricultural production to increase 50% by 2030. The planet's water resources are also under pressure. As Pamela Ronald highlights in her accompanying Essay, the amount of fresh water available per person has decreased 4-fold in the last 60 years and of the water that is available, ~70% is already used for agriculture. Thus, agricultural production must be intensified to feed more people with less water on the same amount of land (given that little undeveloped arable land remains and what does is being lost to urbanization, desertification, and environmental damage). Furthermore, pathogens that cause devastating crop losses continue to spread in the face of increased global commerce and climate change. Given these challenges, there is a pressing need for plant research to produce solutions to ensure food security in a sustainable and safe way. The need is acute in both developed countries and in the less developed parts of the world, where many people endure chronic malnutrition and suffer the long term consequences on their health and well being. Plant scientists, therefore, urgently need to increase the productivity, pathogen resistance, and sustainability of existing crops, and are challenged to domesticate new crops.
Readers of PlantVillage who visited popular pages like the one on tomato late blight would have come across the term “oomycetes” and probably wondered what in the world is an oomycete? It’s the taxon of microbes that groups many plant pathogens such as Phytophthora, Pythium, and the downy mildews. These are destructive pathogens of plants. Phytophthora, which stems from Greek words meaning plant-destroyer, is a diverse group of plant pathogens with over 100 species known to science . It includes the infamous Irish potato famine pathogen Phytophthora infestans. When this pathogen reached Ireland in the 1840s, it triggered famine and mayhem with one million people dead and another million forced to leave the island. Today, the late blight disease caused by P. infestans threatens not only tomatoes and potatoes in your gardens but also commercial and subsistence farming worldwide. Matt Fisher, Sarah Gurr and their colleagues recently estimated that losses due to late blight add up to enough calories to feed hundreds of millions of people.
So what are these oomycetes that are so feared by gardeners and farmers alike? Traditionally oomycetes were thought to be fungi (yeasts, molds and mushrooms). They are not. Modern methods of evolutionary analyses, known as phylogenetics, have cemented the view that oomycetes are only distant relatives of the fungi. In fact, fungi are more closely related to you and I than they are to the oomycetes. Oomycete biologists like to quip “bats are not birds, dolphins are not fish, and oomycetes are not fungi.” In fact, oomycetes turned out to have unexpected marine cousins in brown algae (kelp) and diatoms in a grouping known as the heterokonts. Oomycetes form a very deep branch in the tree of life and may have evolved from marine parasitic microorganisms. Just a few months ago, while I was visiting Christine Strullu-Derrien and Paul Kenrick at the Natural History Museum in London, I had the amazing opportunity to hold a fossil oomycete that is 300 million year old. Already in those ancient times, oomycetes were successful colonizers of plants and may even have been parasitic.
But evolution is a complicated process. More often than widely assumed, it proceeded as a reticulate network rather than a straight line. One example is the transfer of genes from one organism to another. This process, known as horizontal or lateral gene transfer, has occurred frequently in bacteria but is not as well documented in more complex organisms like oomycetes and fungi. Nonetheless, Tom Richard and colleagues at Exeter University reported that oomycetes have at some point in their evolution acquired genes from fungi. Whether this took place hundreds million years ago or more recently is not yet resolved. But as Tom likes to say “oomycetes are 99% not fungi”. How this phenomenon has contributed to the evolution of oomycetes into destructive plant pathogens is an interesting research topic.
But why all the misery? Why are oomycetes the scourge of farmers worldwide? The truth is, although Phytophthora are astonishing plant killers that can wipe out crops in days, the secret of their success is their ability to rapidly adapt to resistant plant varieties. Just like the constantly morphing flu virus, the potato blight pathogen and its relatives continuously spawn new races adapted to the resistant varieties released by plant breeders and even occasionally to new host plants. Like Lewis Carroll’s fictional Red Queen, plant breeders and biotechnologists only hope is to strenuously run to keep in the same place. If only we could produce resistant varieties more often then perhaps we’ll have a chance to outrace the ever-evolving blight pathogen.
So while you lament your blighted potatoes and your dying tomatoes, take a moment to ponder over the awesome parasite that’s making your vegetable garden look so gloomy. That microbe has already colonized plants way before humans emerged on earth. And for hundreds of millions of years it has kept on changing, evolving, and adapting ensuring its uninterrupted survival on an astonishing array of plant species and varieties. When humans domesticated plants, it could not resist the offering and adopted the crops as its new hosts. Ultimately, it moved to new continents and farmlands causing misery and despair. But we haven’t given up. Plant pathologists are hard at work learning more about these parasites and applying new knowledge and technologies to build disease-resistant crops.
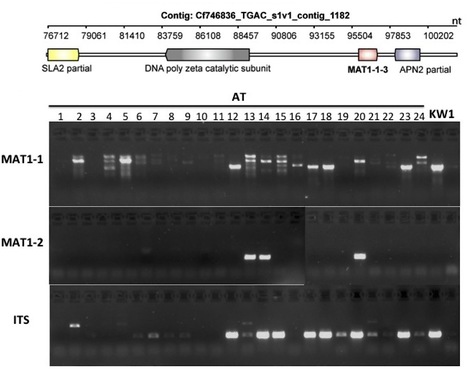
Ash dieback is a fungal disease of ash trees caused by Hymenoscyphus pseudoalbidus that has swept across Europe in the last two decades and is a significant threat to the ash population. This emergent pathogen has been relatively poorly studied and little is known about its genetic make-up. In response to the arrival of this dangerous pathogen in the UK we took the unusual step of providing an open access database and initial sequence datasets to the scientific community for analysis prior to performing an analysis of our own. Our goal was to crowdsource genomic and other analyses and create a community analysing this pathogen. In this report on the evolution of the community and data and analysis obtained in the first year of this activity, we describe the nature and the volume of the contributions and reveal some preliminary insights into the genome and biology of H. pseudoalbidus that emerged. In particular our nascent community generated a first-pass genome assembly containing abundant collapsed AT-rich repeats indicating a typically complex genome structure. Our open science and crowdsourcing effort has brought a wealth of new knowledge about this emergent pathogen within a short time-frame. Our community endeavour highlights the positive impact that open, collaborative approaches can have on fast, responsive modern science.
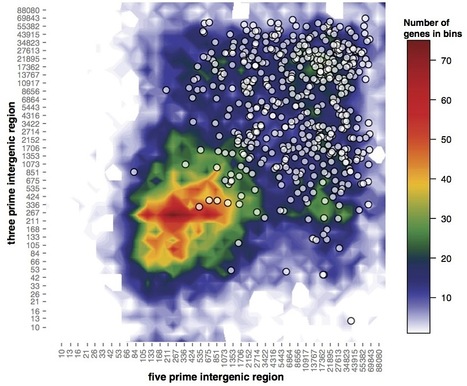
Genome architecture often reflects an organism’s lifestyle and can therefore provide insights into gene function, regulation, and adaptation. In several lineages of plant pathogenic fungi and oomycetes, characteristic repeat-rich and gene-sparse regions harbor pathogenicity-related genes such as effectors. In these pathogens, analysis of genome architecture has assisted the mining for novel candidate effector genes and investigations into patterns of gene regulation and evolution at the whole genome level. Here we describe a two-dimensional data binning method in R with a heatmap-style graphical output to facilitate analysis and visualization of whole genome architecture. The method is flexible, combining whole genome architecture heatmaps with scatter plots of the genomic environment of selected gene sets. This enables analysis of specific values associated with genes such as gene expression and sequence polymorphisms, according to genome architecture. This method enables the investigation of whole genome architecture and reveals local properties of genomic neighborhoods in a clear and concise manner.
Rust fungi cause serious yield reductions on crops, including wheat, barley, soybean, coffee, and represent real threats to global food security. Of these fungi, the flax rust pathogen Melampsora lini has been developed extensively over the past 80 years as a model to understand the molecular mechanisms that underpin pathogenesis. During infection, M. lini secretes virulence effectors to promote disease. The number of these effectors, their function and their degree of conservation across rust fungal species is unknown. To assess this, we sequenced and assembled de novo the genome of M. lini isolate CH5 into 21,130 scaffolds spanning 189 Mbp (scaffold N50 of 31 kbp). Global analysis of the DNA sequence revealed that repetitive elements, primarily retrotransposons, make up at least 45% of the genome. Using ab initio predictions, transcriptome data and homology searches, we identified 16,271 putative protein-coding genes. An analysis pipeline was then implemented to predict the effector complement of M. lini and compare it to that of the poplar rust, wheat stem rust and wheat stripe rust pathogens to identify conserved and species-specific effector candidates. Previous knowledge of four cloned M. lini avirulence effector proteins and two basidiomycete effectors was used to optimise parameters of the effector prediction pipeline. Markov clustering based on sequence similarity was performed to group effector candidates from all four rust pathogens. Clusters containing at least one member from M. lini were further analysed and prioritized based on features including expression in isolated haustoria and infected leaf tissue and conservation across rust species. Herein, we describe 200 of 940 clusters that ranked highest on our priority list, representing 725 flax rust candidate effectors. Our findings on this important model rust species provide insight into how effectors of rust fungi are conserved across species and how they may act to promote infection on their hosts.
Via Francis Martin, Kamoun Lab @ TSL
Fungal and oomycete plant parasites are among the most devastating pathogens of food crops. These microbes secrete effector proteins inside plant cells to manipulate host processes and facilitate colonization. How these effectors reach the host cytoplasm remains an unclear and debated area of plant research. In this article, we examine recent conflicting findings that have generated discussion in the field. We also highlight promising approaches based on studies of both parasite and host during infection. Ultimately, this knowledge may inform future broad spectrum strategies for protecting crops from such pathogens.
Nuclei of arbuscular endomycorrhizal fungi have been described as highly diverse due to their asexual nature and absence of a single cell stage with only one nucleus. This has raised fundamental questions concerning speciation, selection and transmission of the genetic make-up to next generations. Although this concept has become textbook knowledge, it is only based on studying a few loci, including 45S rDNA. To provide a more comprehensive insight into the genetic makeup of arbuscular endomycorrhizal fungi, we applied de novo genome sequencing of individual nuclei of Rhizophagus irregularis. This revealed a surprisingly low level of polymorphism between nuclei. In contrast, within a nucleus, the 45S rDNA repeat unit turned out to be highly diverged. This finding demystifies a long-lasting hypothesis on the complex genetic makeup of arbuscular endomycorrhizal fungi. Subsequent genome assembly resulted in the first draft reference genome sequence of an arbuscular endomycorrhizal fungus. Its length is 141 Mbps, representing over 27,000 protein-coding gene models. We used the genomic sequence to reinvestigate the phylogenetic relationships of Rhizophagus irregulariswith other fungal phyla. This unambiguously demonstrated that Glomeromycota are more closely related to Mucoromycotina than to its postulated sister Dikarya.
Accurate identification of DNA polymorphisms using next-generation sequencing technology is challenging because of a high rate of sequencing error and incorrect mapping of reads to reference genomes. Currently available short read aligners and DNA variant callers suffer from these problems. We developed the Coval software to improve the quality of short read alignments. Coval is designed to minimize the incidence of spurious alignment of short reads, by filtering mismatched reads that remained in alignments after local realignment and error correction of mismatched reads. The error correction is executed based on the base quality and allele frequency at the non-reference positions for an individual or pooled sample. We demonstrated the utility of Coval by applying it to simulated genomes and experimentally obtained short-read data of rice, nematode, and mouse. Moreover, we found an unexpectedly large number of incorrectly mapped reads in ‘targeted’ alignments, where the whole genome sequencing reads had been aligned to a local genomic segment, and showed that Coval effectively eliminated such spurious alignments. We conclude that Coval significantly improves the quality of short-read sequence alignments, thereby increasing the calling accuracy of currently available tools for SNP and indel identification. Coval is available at http://sourceforge.net/projects/coval105/.
Puccinia monoica is a spectacular plant parasitic rust fungus that triggers the formation of flower-like structures (pseudoflowers) in its Brassicaceae host plant Boechera stricta. Pseudoflowers mimic in shape, color, nectar and scent co-occurring and unrelated flowers such as buttercups. They act to attract insects thereby aiding spore dispersal and sexual reproduction of the rust fungus. Although much ecological research has been performed on P.monoica-induced pseudoflowers, this system has yet to be investigated at the molecular or genomic level. To date, the molecular alterations underlying the development of pseudoflowers and the genes involved have not been described. To address this, we performed gene expression profiling to reveal 256 plant biological processes that are significantly altered in pseudoflowers. Among these biological processes, plant genes involved in cell fate specification, regulation of transcription, reproduction, floral organ development, anthocyanin (major floral pigments) and terpenoid biosynthesis (major floral volatile compounds) were down-regulated in pseudoflowers. In contrast, plant genes involved in shoot, cotyledon and leaf development, carbohydrate transport, wax biosynthesis, cutin transport and L-phenylalanine metabolism (pathway that results in phenylethanol and phenylacetaldehyde volatile production) were up-regulated. These findings point to an extensive reprogramming of host genes by the rust pathogen to induce floral mimicry. We also highlight 31 differentially regulated plant genes that are enriched in the biological processes mentioned above, and are potentially involved in the formation of pseudoflowers. This work illustrates the complex perturbations induced by rust pathogens in their host plants, and provides a starting point for understanding the molecular mechanisms of pathogen-induced floral mimicry.
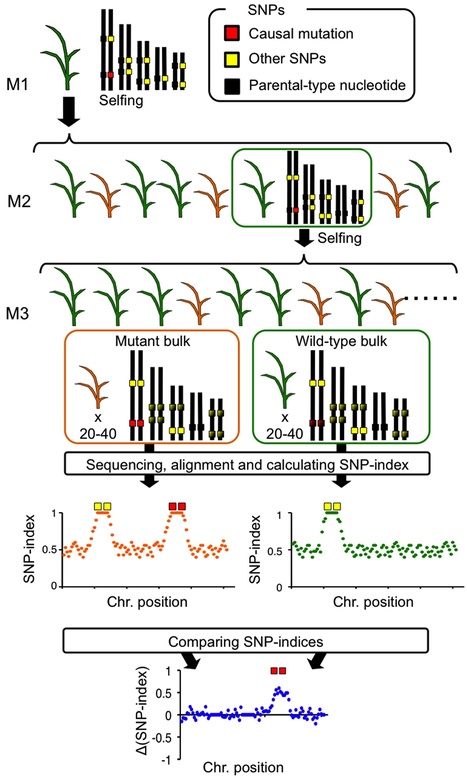
Advances in genome sequencing technologies have enabled researchers and breeders to rapidly associate phenotypic variation to genome sequence differences. We recently took advantage of next-generation sequencing technology to develop MutMap, a method that allows rapid identification of causal nucleotide changes of rice mutants by whole genome resequencing of pooled DNA of mutant F2 progeny derived from crosses made between candidate mutants and the parental line. Here we describe MutMap+, a versatile extension of MutMap, that identifies causal mutations by comparing SNP frequencies of bulked DNA of mutant and wild-type progeny of M3 generation derived from selfing of an M2 heterozygous individual. Notably, MutMap+ does not necessitate artificial crossing between mutants and the wild-type parental line. This method is therefore suitable for identifying mutations that cause early development lethality, sterility, or generally hamper crossing. Furthermore, MutMap+ is potentially useful for gene isolation in crops that are recalcitrant to artificial crosses.
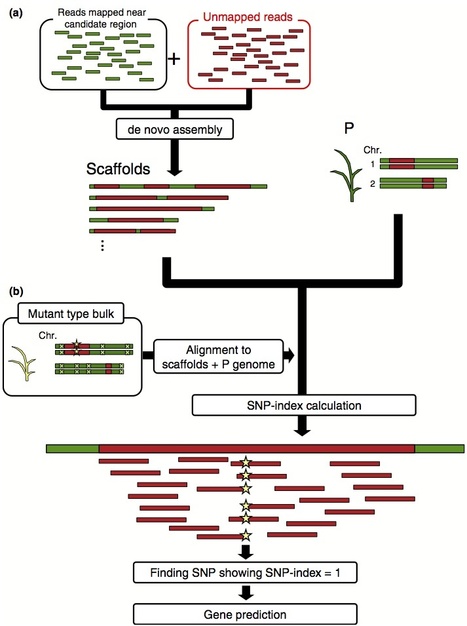
Next-generation sequencing allows the identification of mutations responsible for mutant phenotypes by whole-genome resequencing and alignment to a reference genome. However, when the resequenced cultivar/line displays significant structural variation from the reference genome, mutations in the genome regions missing from the reference (gaps) cannot be identified by simple alignment.Here we report on a method called ‘MutMap-Gap’, which involves delineating a candidate region harboring a mutation of interest using the recently reported MutMap method, followed by de novo assembly, alignment, and identification of the mutation within genome gaps.We applied MutMap-Gap to isolate the blast resistant gene Pii from the rice cv Hitomebore using mutant lines that have lost Pii function.MutMap-Gap should prove useful for cloning genes that exhibit significant structural variations such as disease resistance genes of the nucleotide-binding site-leucine rich repeat (NBS-LRR) class.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...

































Pepper pathogen can handle the 'heat'