Your new post is loading...
 Your new post is loading...
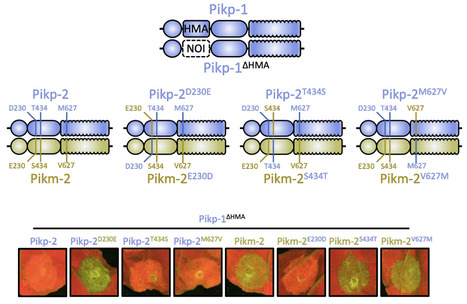
Engineering the plant immune system offers genetic solutions to mitigate crop diseases caused by diverse agriculturally significant pathogens and pests. Modification of intracellular plant immune receptors of the nucleotide-binding leucine rich repeat (NLR) superfamily for expanded recognition of pathogen virulence proteins (effectors) is a promising approach for engineering disease resistance. However, engineering can cause NLR autoactivation, resulting in constitutive defence responses that are deleterious to the plant. This may be due to plant NLRs associating in highly complex signalling networks that co-evolve together, and changes through breeding or genetic modification can generate incompatible combinations, resulting in autoimmune phenotypes. The sensor and helper NLRs of the rice (Oryza sativa) NLR pair Pik have co-evolved, and mismatching between non-co-evolved alleles triggers constitutive activation and cell death. This limits the extent to which protein modifications can be used to engineer pathogen recognition and enhance disease resistance mediated by these NLRs. Here, we dissected incompatibility determinants in the Pik pair in Nicotiana benthamiana and found that heavy metal-associated (HMA) domains integrated in Pik-1 not only evolved to bind pathogen effectors but also likely co-evolved with other NLR domains to maintain immune homeostasis. This explains why changes in integrated domains can lead to autoactivation. We then used this knowledge to facilitate engineering of new effector recognition specificities, overcoming initial autoimmune penalties. We show that by mismatching alleles of the rice sensor and helper NLRs Pik-1 and Pik-2, we can enable the integration of synthetic domains with novel and enhanced recognition specificities. Taken together, our results reveal a strategy for engineering NLRs, which has the potential to allow an expanded set of integrations and therefore new disease resistance specificities in plants.
Via The Sainsbury Lab
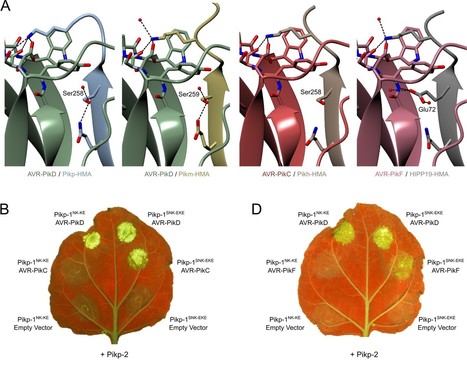
A subset of plant intracellular NLR immune receptors detect effector proteins, secreted by phytopathogens to promote infection, through unconventional integrated domains which resemble the effector’s host targets. Direct binding of effectors to these integrated domains activates plant defenses. The rice NLR receptor Pik-1 binds the Magnaporthe oryzae effector AVR-Pik through an integrated heavy metal-associated (HMA) domain. However, the stealthy alleles AVR-PikC and AVR-PikF avoid interaction with Pik-HMA and evade host defenses. Here, we exploited knowledge of the biochemical interactions between AVR-Pik and its host target, OsHIPP19, to engineer novel Pik-1 variants that respond to AVR-PikC/F. First, we exchanged the HMA domain of Pikp-1 for OsHIPP19-HMA, demonstrating that effector targets can be incorporated into NLR receptors to provide novel recognition profiles. Second, we used the structure of OsHIPP19-HMA to guide the mutagenesis of Pikp-HMA to expand its recognition profile. We demonstrate that the extended recognition profiles of engineered Pikp-1 variants correlate with effector binding in planta and in vitro, and with the gain of new contacts across the effector/HMA interface. Crucially, transgenic rice producing the engineered Pikp-1 variants was resistant to blast fungus isolates carrying AVR-PikC or AVR-PikF. These results demonstrate that effector target-guided engineering of NLR receptors can provide new-to-nature disease resistance in crops.
Via The Sainsbury Lab
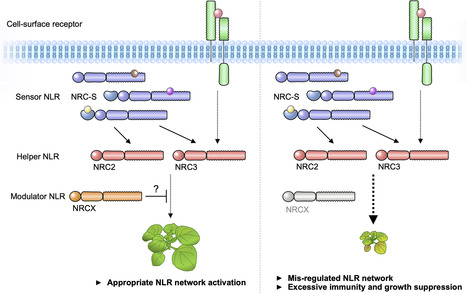
The NRC immune receptor network has evolved in asterid plants from a pair of linked genes into a genetically dispersed and phylogenetically structured network of sensor and helper NLR (nucleotide-binding domain and leucine-rich repeat-containing) proteins. In some species, such as the model plant Nicotiana benthamiana and other Solanaceae, the NRC (NLR-REQUIRED FOR CELL DEATH) network forms up to half of the NLRome, and NRCs are scattered throughout the genome in gene clusters of varying complexities. Here, we describe NRCX, an atypical member of the NRC family that lacks canonical features of these NLR helper proteins, such as a functional N-terminal MADA motif and the capacity to trigger autoimmunity. In contrast to other NRCs, systemic gene silencing of NRCX in N. benthamiana markedly impairs plant growth resulting in a dwarf phenotype. Remarkably, dwarfism of NRCX silenced plants is partially dependent on NRCX paralogs NRC2 and NRC3, but not NRC4. Despite its negative impact on plant growth when silenced systemically, spot gene silencing of NRCX in mature N. benthamiana leaves doesn't result in visible cell death phenotypes. However, alteration of NRCX expression modulates the hypersensitive response mediated by NRC2 and NRC3 in a manner consistent with a negative role for NRCX in the NRC network. We conclude that NRCX is an atypical member of the NRC network that has evolved to contribute to the homeostasis of this genetically unlinked NLR network.
Via The Sainsbury Lab
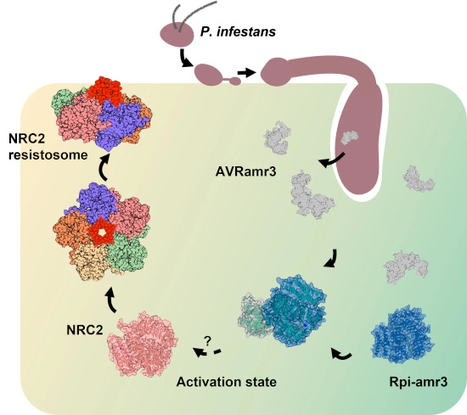
Plant pathogens compromise crop yields. Plants have evolved robust innate immunity that depends in part on intracellular Nucleotide-binding, Leucine rich-Repeat (NLR) immune receptors that activate defense responses upon detection of pathogen-derived effectors. Most “sensor” NLRs that detect effectors require the activity of “helper” NLRs, but how helper NLRs support sensor NLR function is poorly understood. Many Solanaceae NLRs require NRC (NLR-Required for Cell death) class of helper NLRs. We show here that Rpi-amr3, a sensor NLR from Solanum americanum, detects AVRamr3 from the potato late blight pathogen, Phytophthora infestans, and activates oligomerization of helper NLRs NRC2 and NRC4 into high-molecular-weight resistosomes. In contrast, recognition of P. infestans effector AVRamr1 by another sensor NLR Rpi-amr1 induces formation of only the NRC2 resistosome. The activated NRC2 oligomer becomes enriched in membrane fractions. ATP-binding motifs of both Rpi-amr3 and NRC2 are required for NRC2 resistosome formation, but not for the interaction of Rpi-amr3 with its cognate effector. NRC2 resistosome can be activated by Rpi-amr3 upon detection of AVRamr3 homologs from other Phytophthora species. Mechanistic understanding of NRC resistosome formation will underpin engineering crops with durable disease resistance.
Via The Sainsbury Lab
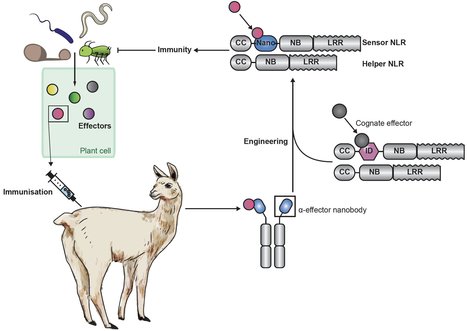
Plant pathogens cause recurrent epidemics, threatening crop yield and global food security. Efforts to retool the plant immune system have been limited to modifying natural components and can be nullified by the emergence of new pathogen strains. Made-to-order synthetic plant immune receptors provide an opportunity to tailor resistance to pathogen genotypes present in the field. In this work, we show that plant nucleotide-binding, leucine-rich repeat immune receptors (NLRs) can be used as scaffolds for nanobody (single-domain antibody fragment) fusions that bind fluorescent proteins (FPs). These fusions trigger immune responses in the presence of the corresponding FP and confer resistance against plant viruses expressing FPs. Because nanobodies can be raised against most molecules, immune receptor-nanobody fusions have the potential to generate resistance against plant pathogens and pests delivering effectors inside host cells.
Via The Sainsbury Lab
Publishing your poster in a DOI repository like Zenodo extends the lifespan of your poster beyond the conference, reaching audiences who would not have seen your poster otherwise and providing a verifiable, citable record of your scholarly output. Open science in Providence This week finds me in Providence, Rhode Island, USA, attending the 19th Congress of IS-MPMI — the International Society for Molecular Plant-Microbe Interactions. This society has been my scientific home throughout my career, and its Congress is renowned for announcements of groundbreaking research in our fast-paced and diverse field. This year, the program is more inclusive than ever, featuring increased representation from the international community and a wide variety of workshops and sessions aimed at career advancement. Among these, a Concurrent Session deserves highlighting. The brainchild of Toulouse-based PhD student Karima El Mahboubi, who is a passionate advocate of open science and a proponent of reforming the publishing system, this session boasts an inspiring array of talks. Notably, Karima managed to secure Jessica Polka, the Executive Director of ASAPbio and a leading voice in the open science movement, as a speaker.
“I believe empathy is the most essential quality of civilization.” — Roger Ebert This post was inspired by behavior I witnessed a few days ago at the conference hotel I was staying at. You know those moments when you courteously wait for people to get on the elevator, only to have them cut ahead of you at the busy checkout counter. Reflecting on this incident, I thought again about the importance of small gestures in our daily interactions. It reminded me of the significance of expressing gratitude, thinking about the people around us and the need to avoid unnecessary rushes.
In the unending coevolutionary dance between plants and microbes, each player impacts the evolution of the other. This article provides an overview of the burgeoning field of Evolutionary Molecular Plant-Microbe Interactions (EVO-MPMI), tracing its progression from foundational science to practical implementation. It delves into central concepts and methodologies, and offers a snapshot of current research. Moreover, the review sheds light on the practical applications of EVO-MPMI, particularly within the realm of disease control. Looking ahead, we discuss potential future trajectories for EVO-MPMI research, spotlighting the innovative tools and technologies propelling the discipline forward. FIRST PUBLISHED AS: Schornack, Sebastian and Kamoun, Sophien. (2023). EVO-MPMI: From fundamental science to practical applications. Zenodo https://doi.org/10.5281/zenodo.8030196
“Please enjoy the journey” — let’s etch these words onto the walls of our laboratories. I have always been a sports enthusiast. There’s something captivating and inspiring about it, although it’s difficult to put into words. Perhaps it’s the incredible athletes and their awe-inspiring feats, or maybe it’s the way sports mirrors the dramas of life itself. I frequently find myself drawing parallels between sports and academia. Take, for example, Arsène Wenger, the studious Frenchman who coached Arsenal to six trophies and is known for having modernized English football. He was famously referred to as “The Professor” or, perhaps jokingly, “Le Professeur” during the pinnacle of his career. That title even became the name of his biography. Regrettably, similar to many professors, Wenger ended his career perceived as dogmatic and out-of-touch, living off past glories. This serves as a valuable lesson for those of us embarking on the later stages of our own academic journeys.
A visit to Halle, 25 years later, and a workshop with early career researchers brought into focus what should be the primary mission of scientists.
Rawit, a Thai student, was awarded a scholarship to spend a year studying at Oxford University. Things didn’t go to plan. Rawit Longsaward is a Ph.D. student at Mahidol University in Thailand. In 2021/2022, he was awarded a scholarship to spend a year in the Plant Chemetics Laboratory of Renier van der Hoorn at the University of Oxford. His objective was to study the extensively researched ribonuclease activity of plant defense-induced proteins belonging to the PR-10 family. Things didn’t go to plan, and his account of the events is detailed below. It is disheartening to note that his experience – wasting time and money due to irreproducible published results – is not unique.
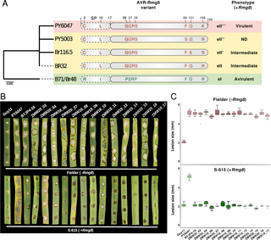
Wheat, one of the most important food crops, is threatened by a blast disease pandemic. Here, we show that a clonal lineage of the wheat blast fungus recently spread to Asia and Africa following two independent introductions from South America. Through a combination of genome analyses and laboratory experiments, we show that the decade-old blast pandemic lineage can be controlled by the Rmg8 disease resistance gene and is sensitive to strobilurin fungicides. However, we also highlight the potential of the pandemic clone to evolve fungicide-insensitive variants and sexually recombine with African lineages. This underscores the urgent need for genomic surveillance to track and mitigate the spread of wheat blast outside of South America and to guide preemptive wheat breeding for blast resistance.
Via The Sainsbury Lab
Expatriate scientists have an important role to play in advancing science and helping their colleagues back home. This post is to encourage you to do something. Anything. Ya rayah (Arabic: يا رايح)
Many scientists work abroad, often in Western countries, joining a growing number of expatriates from their home countries. The struggles and sacrifices faced by these individuals are portrayed in the popular Algerian ballad “Ya Rayah,” meaning “The Traveler” or “The Exiled.” Originally written and performed by the Algerian musician Dahmane El Harrachi in the 1970s, my personal favorite version is the cover by the late Algerian rock star Rachid Taha. Taha’s “gruff voice” — as described by the New York Times in his 2018 obituary — combined with his own activism for immigrant rights, lend a deep emotional weight to the song.
|
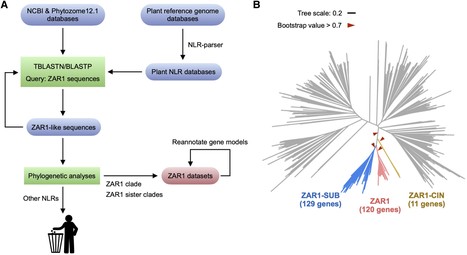
Plant nucleotide-binding leucine-rich repeat (NLR) immune receptors generally exhibit hallmarks of rapid evolution, even at the intraspecific level. We used iterative sequence similarity searches coupled with phylogenetic analyses to reconstruct the evolutionary history of HOPZ-ACTIVATED RESISTANCE1 (ZAR1), an atypically conserved NLR that traces its origin to early flowering plant lineages ∼220 to 150 million yrs ago (Jurassic period). We discovered 120 ZAR1 orthologs in 88 species, including the monocot Colocasia esculenta, the magnoliid Cinnamomum micranthum, and most eudicots, notably the Ranunculales species Aquilegia coerulea, which is outside the core eudicots. Ortholog sequence analyses revealed highly conserved features of ZAR1, including regions for pathogen effector recognition and cell death activation. We functionally reconstructed the cell death activity of ZAR1 and its partner receptor-like cytoplasmic kinase (RLCK) from distantly related plant species, experimentally validating the hypothesis that ZAR1 evolved to partner with RLCKs early in its evolution. In addition, ZAR1 acquired novel molecular features. In cassava (Manihot esculenta) and cotton (Gossypium spp.), ZAR1 carries a C-terminal thioredoxin-like domain, and in several taxa, ZAR1 duplicated into 2 paralog families, which underwent distinct evolutionary paths. ZAR1 stands out among angiosperm NLR genes for having experienced relatively limited duplication and expansion throughout its deep evolutionary history. Nonetheless, ZAR1 also gave rise to noncanonical NLRs with integrated domains and degenerated molecular features.
Via The Sainsbury Lab
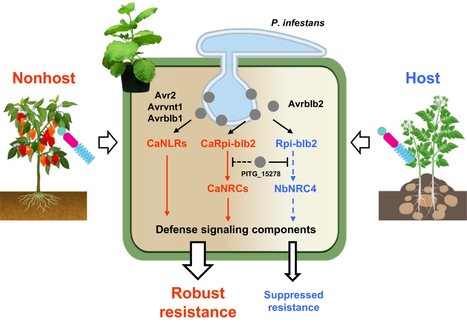
Nonhost resistance (NHR) is a robust plant immune response against non-adapted pathogens. A number of nucleotide-binding leucine-rich repeat (NLR) proteins that recognize non-adapted pathogens have been identified, although the underlying molecular mechanisms driving robustness of NHR are still unknown. Here, we screened 57 effectors of the potato late blight pathogen Phytophthora infestans in nonhost pepper (Capsicum annuum) to identify avirulence effector candidates. Selected effectors were tested against 436 genome-wide cloned pepper NLRs, and we identified multiple functional NLRs that recognize P. infestans effectors and confer disease resistance in the Nicotiana benthamiana as a surrogate system. The identified NLRs were homologous to known NLRs derived from wild potatoes that recognize P. infestans effectors such as Avr2, Avrblb1, Avrblb2, and Avrvnt1. The identified CaRpi-blb2 is a homologue of Rpi-blb2, recognizes Avrblb2 family effectors, exhibits feature of lineage-specifically evolved gene in microsynteny and phylogenetic analyses, and requires pepper-specific NRC (NLR required for cell death)-type helper NLR for proper function. Moreover, CaRpi-blb2–mediated hypersensitive response and blight resistance were more tolerant to suppression by the PITG_15 278 than those mediated by Rpi-blb2. Combined results indicate that pepper has stacked multiple NLRs recognizing effectors of non-adapted P. infestans, and these NLRs could be more tolerant to pathogen-mediated immune suppression than NLRs derived from the host plants. Our study suggests that NLRs derived from nonhost plants have potential as untapped resources to develop crops with durable resistance against fast-evolving pathogens by stacking the network of nonhost NLRs into susceptible host plants.
Via The Sainsbury Lab
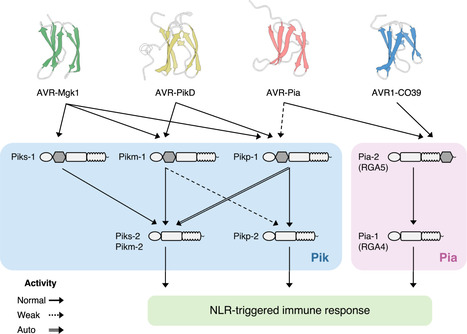
Studies focused solely on single organisms can fail to identify the networks underlying host–pathogen gene-for-gene interactions. Here, we integrate genetic analyses of rice (Oryza sativa, host) and rice blast fungus (Magnaporthe oryzae, pathogen) and uncover a new pathogen recognition specificity of the rice nucleotide-binding domain and leucine-rich repeat protein (NLR) immune receptor Pik, which mediates resistance to M. oryzae expressing the avirulence effector gene AVR-Pik. Rice Piks-1, encoded by an allele of Pik-1, recognizes a previously unidentified effector encoded by the M. oryzae avirulence gene AVR-Mgk1, which is found on a mini-chromosome. AVR-Mgk1 has no sequence similarity to known AVR-Pik effectors and is prone to deletion from the mini-chromosome mediated by repeated Inago2 retrotransposon sequences. AVR-Mgk1 is detected by Piks-1 and by other Pik-1 alleles known to recognize AVR-Pik effectors; recognition is mediated by AVR-Mgk1 binding to the integrated heavy metal-associated (HMA) domain of Piks-1 and other Pik-1 alleles. Our findings highlight how complex gene-for-gene interaction networks can be disentangled by applying forward genetics approaches simultaneously to the host and pathogen. We demonstrate dynamic coevolution between an NLR integrated domain and multiple families of effector proteins.
Via The Sainsbury Lab
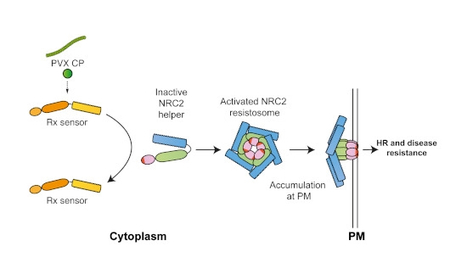
Nucleotide-binding domain leucine-rich repeat (NLR) immune receptors are important components of plant and metazoan innate immunity that can function as individual units or as pairs or networks. Upon activation, NLRs form multiprotein complexes termed resistosomes or inflammasomes. Although metazoan paired NLRs, such as NAIP/NLRC4, form hetero-complexes upon activation, the molecular mechanisms underpinning activation of plant paired NLRs, especially whether they associate in resistosome hetero-complexes, is unknown. In asterid plant species, the NLR required for cell death (NRC) immune receptor network is composed of multiple resistance protein sensors and downstream helpers that confer immunity against diverse plant pathogens. Here, we show that pathogen effector-activation of the NLR proteins Rx (confers virus resistance), and Bs2 (confers bacterial resistance) leads to oligomerization of their helper NLR, NRC2. Activated Rx does not oligomerize or enter into a stable complex with the NRC2 oligomer and remains cytoplasmic. In contrast, activated NRC2 oligomers accumulate in membrane-associated puncta. We propose an activation-and-release model for NLRs in the NRC immune receptor network. This points to a distinct activation model compared with mammalian paired NLRs.
Via The Sainsbury Lab
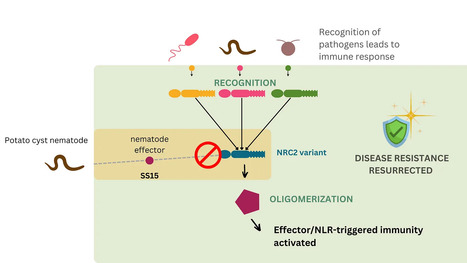
Parasites counteract host immunity by suppressing helper nucleotide binding and leucine-rich repeat (NLR) proteins that function as central nodes in immune receptor networks. Understanding the mechanisms of immunosuppression can lead to strategies for bioengineering disease resistance. Here, we show that a cyst nematode virulence effector binds and inhibits oligomerization of the helper NLR protein NRC2 by physically preventing intramolecular rearrangements required for activation. An amino acid polymorphism at the binding interface between NRC2 and the inhibitor is sufficient for this helper NLR to evade immune suppression, thereby restoring the activity of multiple disease resistance genes. This points to a potential strategy for resurrecting disease resistance in crop genomes.
There’s a bright future for bioengineering of plant immune receptors, a future that’s deeply rooted in fundamental research and discovery. This article is available on a CC-BY license via Zenodo. Cite as: Kamoun, S. (2023) How to trick a plant pathogen: A bright future for bioengineering of plant immune receptors. https://doi.org/10.5281/zenodo.8127512
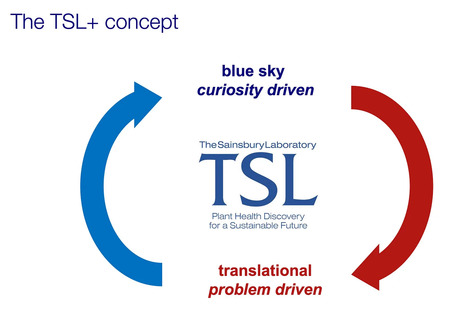
At The Sainsbury Lab, merging fundamental and applied research has led to unexpected discoveries and real-world impacts. The “Gained in Translation” concept highlights the value of open-ended, problem-solving research. This week, as I attended #TRIC23 the ‘Translational Research in Crops’ conference organized by VIB in Ghent, Belgium, I was reminded of the “Gained in Translation” concept that has shaped our research ethos at The Sainsbury Lab (TSL). Roughly a decade ago, we launched the TSL+ concept. This initiative encourages lab leaders to move beyond pure curiosity-driven research and to delve into problem-solving, application-oriented studies. One of my slides encapsulating this philosophy sparked intriguing conversations at the conference. Today, it’s fair to say that this merger of fundamental and applied research under a single roof has been a resounding success. It has not only enriched our research portfolio but also broadened the range of our discoveries. --- This article is available on a CC-BY license via Zenodo. Cite as: Kamoun, S. (2023) Gained in translation: Bridging the gap between curiosity and applied research. https://doi.org/10.5281/zenodo.8074998
The legacy of my great-grandfather Mohamed Said Kamoun lives on. His principles transcend generations and serve as guiding lights for anyone seeking success, whether in business or personal life.
The Daily Express, a UK tabloid, has published a positive article about gene editing (GE). The article highlights the potential benefits of gene editing, including its ability to treat genetic diseases and improve agricultural yields. The piece reflects a growing trend of mainstream media coverage of gene editing, and signals a shift towards more positive and optimistic coverage of this cutting-edge technology that holds so much promise for improving the crops we grow. The Daily Express’ article suggests that the potential benefits of this technology are becoming increasingly recognized and celebrated.
The habit of naming biological tools and material after living organisms is unhelpful and confusing. Let’s stop this. In this time of year, the common dandelion— Taraxacum officinale — can be found almost everywhere. Their bright yellow flowers and fluffy seed heads are a common sight in lawns, gardens, and other green spaces. The mild temperatures and increased rainfall of spring provide ideal growing conditions for dandelions, allowing them to quickly reproduce and spread. Dandelions are often considered a nuisance and are therefore disliked by many people. This is because they are highly invasive and can quickly take over lawns, gardens, and other green spaces. They reproduce rapidly and can spread through both their seeds and their long taproots, making them difficult to control. In addition, the bright yellow flowers of dandelions are often viewed as unsightly, and their fluffy seed heads can quickly spread throughout the surrounding area, further contributing to their invasive nature. I never really understood this dislike of dandelions. They are an important source of nectar and pollen for bees and other pollinators, and their leaves can be used in salads or cooked as a vegetable, as they are rich in vitamins and minerals. Also, they have several amazing biological features that are described in the ~1,000 or so scientific articles about the dandelion genus Taraxacum. The fluffy, white “parachutes” that surround the seeds allow them to be carried great distances and colonize new areas by even the slightest breeze. They have a deep taproot system that can reach up to several meters underground, making them particularly drought-tolerant. They are also able to grow and reproduce at an incredibly fast rate, with some plants producing up to 15,000 seeds in a single growing season.
|



 Your new post is loading...
Your new post is loading...